Amygdala-liver signalling orchestrates glycaemic responses to stress
- PMID: 40903586
- PMCID: PMC12527908
- DOI: 10.1038/s41586-025-09420-1
Amygdala-liver signalling orchestrates glycaemic responses to stress
Abstract
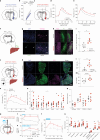
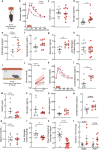
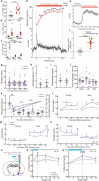
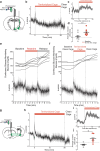
Behavioural adaptations to environmental threats are crucial for survival1,2 and necessitate rapid deployment of energy reserves3-5. The amygdala coordinates behavioural adaptations to threats6, but little is known about its involvement in underpinning metabolic adaptations. Here we show that acute stress activates medial amygdala (MeA) neurons that innervate the ventromedial hypothalamus (MeAVMH neurons), which precipitates hyperglycaemia and hypophagia. The glycaemic actions of MeAVMH neurons occur independently of adrenal or pancreatic glucoregulatory hormones. Using whole-body virus tracing, we identify a polysynaptic connection from MeA to the liver that promotes the rapid synthesis of glucose by hepatic gluconeogenesis. Repeated stress exposure disrupts MeA control of blood glucose, resulting in diabetes-like dysregulation of glucose homeostasis. Our findings reveal an amygdala-liver axis that regulates rapid glycaemic adaptations to stress and links recurrent stress to metabolic dysfunction.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: S.A.S. is a named inventor of patents granted by the United States Patent and Trademark Office: ‘Compositions and Methods to Modulate Cell Activity’ (US 9,399,063 and US 10,064,941) and ‘Ferritin nanoparticle compositions and methods to modulate cell activity’ (US 10,786,570). P.J.K. is a co-founder of Eolas Therapeutics, which is developing novel treatments for substance use disorders. P.J.K. is a consultant for EpiVario.
Figures









Update of
-
Amygdala-liver signaling orchestrates rapid glycemic responses to stress and drives stress-induced metabolic dysfunction.Res Sq [Preprint]. 2024 Mar 29:rs.3.rs-2924278. doi: 10.21203/rs.3.rs-2924278/v1. Res Sq. 2024. Update in: Nature. 2025 Oct;646(8085):697-706. doi: 10.1038/s41586-025-09420-1. PMID: 38585822 Free PMC article. Updated. Preprint.
References
-
- Krishnan, V. et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell131, 391–404 (2007). - PubMed
-
- Mészáros, K., Lang, C. H., Bagby, G. J. & Spitzer, J. J. In vivo glucose utilization by individual tissues during nonlethal hypermetabolic sepsis. FASEB J.2, 3083–3086 (1988). - PubMed
-
- Lang, C. H. & Dobrescu, C. Sepsis-induced increases in glucose uptake by macrophage-rich tissues persist during hypoglycemia. Metabolism40, 585–593 (1991). - PubMed
-
- van der Zwaluw, N. L., van de Rest, O., Kessels, R. P. & de Groot, L. C. Short-term effects of glucose and sucrose on cognitive performance and mood in elderly people. J. Clin. Exp. Neuropsychol.36, 517–527 (2014). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

