Brain-wide representations of prior information in mouse decision-making
- PMID: 40903597
- PMCID: PMC12408363
- DOI: 10.1038/s41586-025-09226-1
Brain-wide representations of prior information in mouse decision-making
Abstract
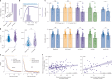
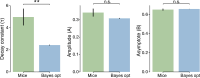
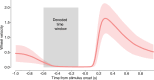
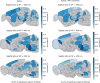
The neural representations of prior information about the state of the world are poorly understood1. Here, to investigate them, we examined brain-wide Neuropixels recordings and widefield calcium imaging collected by the International Brain Laboratory. Mice were trained to indicate the location of a visual grating stimulus, which appeared on the left or right with a prior probability alternating between 0.2 and 0.8 in blocks of variable length. We found that mice estimate this prior probability and thereby improve their decision accuracy. Furthermore, we report that this subjective prior is encoded in at least 20% to 30% of brain regions that, notably, span all levels of processing, from early sensory areas (the lateral geniculate nucleus and primary visual cortex) to motor regions (secondary and primary motor cortex and gigantocellular reticular nucleus) and high-level cortical regions (the dorsal anterior cingulate area and ventrolateral orbitofrontal cortex). This widespread representation of the prior is consistent with a neural model of Bayesian inference involving loops between areas, as opposed to a model in which the prior is incorporated only in decision-making areas. This study offers a brain-wide perspective on prior encoding at cellular resolution, underscoring the importance of using large-scale recordings on a single standardized task.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures










References
-
- Knill, D. C. & Pouget, A. The Bayesian brain: the role of uncertainty in neural coding and computation. Trends Neurosci.27, 712–719 (2004). - PubMed
-
- Ernst, M. O. & Banks, M. S. Humans integrate visual and haptic information in a statistically optimal fashion. Nature415, 429–433 (2002). - PubMed
-
- Jacobs, R. A. Optimal integration of texture and motion cues to depth. Vision Res.39, 3621–3629 (1999). - PubMed
-
- Mamassian, P., Knill, D. C. & Kersten, D. The perception of cast shadows. Trends Cogn. Sci.2, 288–295 (1998). - PubMed
-
- Weiss, Y., Simoncelli, E. P. & Adelson, E. H. Motion illusions as optimal percepts. Nat. Neurosci.5, 598–604 (2002). - PubMed

