Localized high-risk prostate cancer harbors an androgen receptor activity-low subpopulation susceptible to HER2 inhibition
- PMID: 40906535
- PMCID: PMC12618079
- DOI: 10.1172/JCI189900
Localized high-risk prostate cancer harbors an androgen receptor activity-low subpopulation susceptible to HER2 inhibition
Abstract
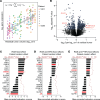
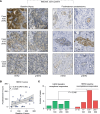
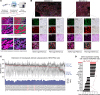
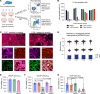
BACKGROUNDLocalized high-risk prostate cancer (PCa) often recurs despite neoadjuvant androgen deprivation therapy (ADT). We sought to identify baseline molecular programs that predict pathologic response and reveal targetable vulnerabilities.METHODSWe profiled 147 biopsy foci from 48 MRI-visible lesions in 37 patients before 6 months of ADT plus enzalutamide and radical prostatectomy. Residual cancer burden (RCB) at prostatectomy was the primary outcome. Analyses incorporated PTEN loss, TMPRSS2:ERG status, and HER2/androgen receptor (AR) immunohistochemistry on baseline and posttreatment tissues. Findings were evaluated in an external transcriptional cohort (n = 121) and by multiplex immunostaining in an independent cohort (n = 61). Functional assays tested enzalutamide-responsive enhancers near ERBB2 and sensitivity to HER2 inhibition.RESULTSA baseline, HER2-associated transcriptional program correlated with higher RCB and inversely with AR activity, independent of PTEN and ERG. Exceptional responders had lower HER2 protein levels in pretreatment biopsy specimens. The inverse AR-HER2 relationship recurred across data sets and multiplex immunostaining, which revealed coexisting AR-high/HER2-low and HER2-high/AR-low subpopulations. Enzalutamide inhibited AR-mediated repression of ERBB2. HER2-high/AR-low cells present before therapy resisted ADT yet were sensitive to HER2 inhibitors; combining HER2 inhibitors with enzalutamide increased tumor cell killing. These findings were reproduced in the external cohort and orthogonal assays.CONCLUSIONBaseline HER2 activity marks intrinsic resistance to neoadjuvant ADT in localized high-risk PCa and identifies a preexisting, targetable AR-low subpopulation. HER2-directed therapy, alone or with AR blockade, warrants clinical evaluation.TRIAL REGISTRATIONClinicalTrials.gov registration: NCT02430480.FUNDINGProstate Cancer Foundation; Department of Defense Prostate Cancer Research Program; National Institutes of Health.
Keywords: Genetics; Molecular pathology; Oncogenes; Oncology; Prostate cancer.
Figures




Update of
-
Localized high-risk prostate cancer harbors an androgen receptor low subpopulation susceptible to HER2 inhibition.medRxiv [Preprint]. 2024 Feb 11:2024.02.09.24302395. doi: 10.1101/2024.02.09.24302395. medRxiv. 2024. Update in: J Clin Invest. 2025 Sep 4;135(22):e189900. doi: 10.1172/JCI189900. PMID: 38370835 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

