Deep Learning Modeling to Differentiate Multiple Sclerosis From MOG Antibody-Associated Disease
- PMID: 40906978
- PMCID: PMC12413738
- DOI: 10.1212/WNL.0000000000214075
Deep Learning Modeling to Differentiate Multiple Sclerosis From MOG Antibody-Associated Disease
Abstract
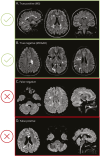
Background and objectives: Multiple sclerosis (MS) is common in adults while myelin oligodendrocyte glycoprotein antibody-associated disease (MOGAD) is rare. Our previous machine-learning algorithm, using clinical variables, ≤6 brain lesions, and no Dawson fingers, achieved 79% accuracy, 78% sensitivity, and 80% specificity in distinguishing MOGAD from MS but lacked validation. The aim of this study was to (1) evaluate the clinical/MRI algorithm for distinguishing MS from MOGAD, (2) develop a deep learning (DL) model, (3) assess the benefit of combining both, and (4) identify key differentiators using probability attention maps (PAMs).
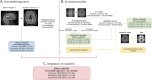
Methods: This multicenter, retrospective, cross-sectional MAGNIMS study included scans from 19 centers. Inclusion criteria were as follows: adults with non-acute MS and MOGAD, with high-quality T2-fluid-attenuated inversion recovery and T1-weighted scans. Brain scans were scored by 2 readers to assess the performance of the clinical/MRI algorithm on the validation data set. A DL-based classifier using a ResNet-10 convolutional neural network was developed and tested on an independent validation data set. PAMs were generated by averaging correctly classified attention maps from both groups, identifying key differentiating regions.
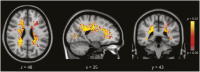
Results: We included 406 MRI scans (218 with relapsing remitting MS [RRMS], mean age: 39 years ±11, 69% F; 188 with MOGAD, age: 41 years ±14, 61% F), split into 2 data sets: a training/testing set (n = 265: 150 with RRMS, age: 39 years ±10, 72% F; 115 with MOGAD, age: 42 years ±13, 61% F) and an independent validation set (n = 141: 68 with RRMS, age: 40 years ±14, 65% F; 73 with MOGAD, age: 40 years ±15, 63% F). The clinical/MRI algorithm predicted RRMS over MOGAD with 75% accuracy (95% CI 67-82), 96% sensitivity (95% CI 88-99), and specificity 56% (95% CI 44-68) in the validation cohort. The DL model achieved 77% accuracy (95% CI 64-89), 73% sensitivity (95% CI 57-89), and 83% specificity (95% CI 65-96) in the training/testing cohort, and 70% accuracy (95% CI 63-77), 67% sensitivity (95% CI 55-79), and 73% specificity (95% CI 61-83) in the validation cohort without retraining. When combined, the classifiers reached 86% accuracy (95% CI 81-92), 84% sensitivity (95% CI 75-92), and 89% specificity (95% CI 81-96). PAMs identified key region volumes: corpus callosum (1872 mm3), left precentral gyrus (341 mm3), right thalamus (193 mm3), and right cingulate cortex (186 mm3) for identifying RRMS and brainstem (629 mm3), hippocampus (234 mm3), and parahippocampal gyrus (147 mm3) for identifying MOGAD.
Discussion: Both classifiers effectively distinguished RRMS from MOGAD. The clinical/MRI model showed higher sensitivity while the DL model offered higher specificity, suggesting complementary roles. Their combination improved diagnostic accuracy, and PAMs revealed distinct damage patterns. Future prospective studies should validate these models in diverse, real-world settings.
Classification of evidence: This study provides Class III evidence that both a clinical/MRI algorithm and an MRI-based DL model accurately distinguish RRMS from MOGAD.
Conflict of interest statement
R. Cortese was awarded a MAGNIMS-ECTRIMS fellowship in 2019; received speaker honoraria/travel support from Roche, Merck Serono, UCB, Sanofi Genzyme, Alexion, Novartis, and Janssen; and received a research grant from the Italian Ministry of University and Research, Research Project of Relevant National Interest (PRIN), project code: 2022PR3PEY. M.P. Amato has served on Scientific Advisory Boards for Biogen, Novartis, Roche, Merck, Sanofi Genzyme, and Teva; received speaker honoraria from Biogen, Merck, Sanofi Genzyme, Roche, Novartis, and Teva; and received research grants for her Institution from Biogen, Merck, Sanofi Genzyme, Novartis, and Roche. G. Arrambide has received speaking honoraria; has received compensation for consulting services or participation in advisory boards from Merck, Roche, and Horizon Therapeutics; has received travel support for scientific meetings from Novartis, Roche, and ECTRIMS; is editor of
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
