Gene actionability according to the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) in No Specific Molecular Profile (NSMP) endometrial cancer
- PMID: 40907210
- PMCID: PMC12446535
- DOI: 10.1016/j.esmoop.2025.105755
Gene actionability according to the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) in No Specific Molecular Profile (NSMP) endometrial cancer
Abstract
Background: The No Specific Molecular Profile (NSMP) subtype accounts for ∼30%-40% of endometrial cancer (EC), comprising a heterogeneous group of EC.
Patients and methods: The primary outcome of this study was the prevalence of actionable genomic alterations in NSMP EC, classified according to the European Society for Medical Oncology (ESMO) Scale for Clinical Actionability of molecular Targets (ESCAT). Oncogenic and likely oncogenic alterations, pathways, and co-mutation patterns were reported. The analysis was stratified by risk group according to the European Society of Gynaecological Oncology (ESGO)-ESMO-European SocieTy for Radiotherapy and Oncology (ESTRO) guidelines. Patients with NSMP EC enrolled in the FPG500 comprehensive cancer genome profiling program (NCT06020625) were included.
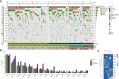
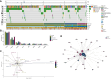
Results: Two hundred and fifty-three patients with NSMP EC of any International Federation of Gynecology and Obstetrics (FIGO) stage were enrolled between 1 January 2022 and 31 December 2023. Median age was 62 years, and the most frequent histotype was endometrioid (97%). Ninety-five percent of patients were estrogen receptor positive. Two hundred and thirty-three patients (92%) had at least one ESCAT tier I-III alteration. The most frequent variants were in PTEN [88%, 95% confidence interval (CI) 84% to 92%], PIK3CA (42%, 95% CI 36% to 49%), FGFR2 (15%, 95% CI 11% to 20%), and AKT1 (6%, 95% CI 3% to 10%); 4% (95% CI 2% to 8%) of patients had an ESR1 variant, while KRAS G12C was found in 3% (95% CI 1% to 6%) of patients. The majority of PTEN variants were on the R130 hotspot. More frequent PIK3CA hotspot variants were H1047R (9%), E545D/K/Q/A (6%), and E542K (4%). In the overall population, PIK3CA with PIK3R1 (odds ratio [OR] = 0.07, P value = 4.25 × 10-14), KRAS with CTNNB1 (OR = 0.03, P value = 1.98 × 10-7), and AKT1 and PTEN (OR = 0.06, P value = 5.73 × 10-6) were mutually exclusive. Significant co-occurrence was found between PTEN and ARID1A (OR = 6.94, P value = 3.39 × 10-6) and PTEN with PIK3R1 (OR = 4.36, P value = 0.009). An alteration in the phosphatidylinositol-3 kinase (PI3K) pathway was found in 94% of patients in the overall population and in 98% of patients with an ESCAT tier I-III alteration.
Conclusion: Our findings highlight potentially actionable alterations in NSMP EC patients, supporting the exploration of tailored molecular-matched therapies according to risk groups.
Keywords: ESCAT; No Specific Molecular Profile endometrial cancer; genomic description; target therapy.
Copyright © 2025 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Disclosure CN declares travel support from MSD, Illumina, Menarini, and AZ; and honoraria from Veeva, GSK, MSD, AZ, Altems, Illumina, and Guardant Health. GS reports research support from MSD and honoraria from Clovis Oncology; and consultant for Tesaro and Johnson & Johnson. FF reports research supports for MSD, GSK, SYSMEX, and STRYKER; honoraria from SYSMEX, MSD, GSK, and STRYKER; and consultant for Clovis Oncology, GSK, MSD, and PharmaMar. All other authors have declared no conflicts of interest.
Figures

References
-
- McConechy M.K., Talhouk A., Leung S., et al. Endometrial carcinomas with POLE exonuclease domain mutations have a favorable prognosis. Clini Cancer Res. 2016;22:2865–2873. - PubMed
-
- Jamieson A., Thompson E.F., Huvila J., Gilks C.B., McAlpine J.N. p53abn Endometrial cancer: understanding the most aggressive endometrial cancers in the era of molecular classification. Int J Gynecol Cancer. 2021;31:907–913. - PubMed
-
- Jamieson A., Huvila J., Chiu D., et al. Grade and estrogen receptor expression identify a subset of no specific molecular profile endometrial carcinomas at a very low risk of disease-specific death. Mod Pathol. 2023;36 - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

