Genetic design of soybean hosts and bradyrhizobial endosymbionts reduces N2O emissions from soybean rhizosphere
- PMID: 40908282
- PMCID: PMC12411642
- DOI: 10.1038/s41467-025-63223-6
Genetic design of soybean hosts and bradyrhizobial endosymbionts reduces N2O emissions from soybean rhizosphere
Abstract
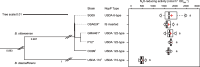
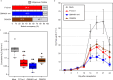
Soybeans fix atmospheric N2 through symbiosis with rhizobia. The relationship between rhizobia and soybeans, particularly those with high nitrous oxide (N2O)-reducing (N2OR) activities, can be leveraged to reduce N2O emissions from agricultural soils. However, inoculating soybeans with these rhizobia under field conditions often fails because of the competition from indigenous rhizobia that possess low or no N2OR activity. In this work, we utilize natural incompatibility systems between soybean and rhizobia to address this challenge. Specifically, Rj2 and GmNNL1 inhibit certain rhizobial infections in response to NopP, an effector protein. By combining a soybean line with a hybrid accumulation of the Rj2 and GmNNL1 genes and bradyrhizobia lacking the nopP gene, we develop a soybean-bradyrhizobial symbiosis system in which strains with high N2OR activity predominantly infect. Our optimize symbiotic system substantially reduces N2O emissions in field and laboratory tests, presenting a promising approach for sustainable agricultural practices.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Tian, H. et al. A comprehensive quantification of global nitrous oxide sources and sinks. Nature586, 248–256 (2020). - PubMed
-
- Galloway, J. N. et al. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science320, 889–892 (2008). - PubMed
-
- Kuypers, M. M. M., Marchant, H. K. & Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol16, 263–276 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

