Next-generation sequencing applications in food science: fundamentals and recent advances
- PMID: 40909218
- PMCID: PMC12405390
- DOI: 10.3389/fbioe.2025.1638957
Next-generation sequencing applications in food science: fundamentals and recent advances
Abstract
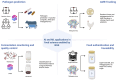
Next-generation sequencing (NGS) has revolutionized food science, offering unprecedented insights into microbial communities, food safety, fermentation, and product authenticity. NGS techniques, including metagenetics, metagenomics, and metatranscriptomics, enable culture-independent pathogen detection, antimicrobial resistance surveillance, and detailed microbial profiling, significantly improving food safety monitoring and outbreak prevention. In food fermentation, NGS has enhanced our understanding of microbial interactions, flavor formation, and metabolic pathways, contributing to optimized starter cultures and improved product quality. Furthermore, NGS has become a valuable tool in food authentication and traceability, ensuring product integrity and detecting fraud. Despite its advantages, challenges such as high sequencing costs, data interpretation complexity, and the need for standardized workflows remain. Future research focusing on optimizing real-time sequencing technologies, expanding multi-omics approaches, and addressing regulatory frameworks is suggested to fully harness NGS's potential in ensuring food safety, quality, and innovation.
Keywords: fermentation; food authentication; food microbiome; food safety; next-generation sequencing.
Copyright © 2025 Tigrero-Vaca, Díaz, Gu and Cevallos-Cevallos.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Abdi G., Tarighat M. A., Jain M., Tendulkar R., Tendulkar M., Barwant M. (2024). “Revolutionizing genomics: exploring the potential of next-generation sequencing,” in Advances in bioinformatics (Singapore: Springer Nature Singapore; ), 1–33. 10.1007/978-981-99-8401-5_1 - DOI
-
- Afifa K., Hossain A., Hossain Md. S., Kamruzzaman Munshi M., Huque R. (2021). Detection of species adulteration in meat products and Mozzarella-type cheeses using duplex PCR of mitochondrial cyt b gene: a food safety concern in Bangladesh. Food Chem. Mol. Sci. 2, 100017. 10.1016/j.fochms.2021.100017 - DOI - PMC - PubMed
-
- Alcock B. P., Huynh W., Chalil R., Smith K. W., Raphenya A. R., Wlodarski M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, D690–D699. 10.1093/nar/gkac920 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

