The genome sequence of the zebra danio, Danio rerio (Hamilton, 1822) (Cypriniformes: Danionidae)
- PMID: 40909362
- PMCID: PMC12405855
- DOI: 10.12688/wellcomeopenres.24569.1
The genome sequence of the zebra danio, Danio rerio (Hamilton, 1822) (Cypriniformes: Danionidae)
Abstract
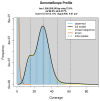
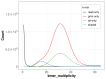
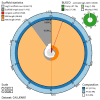
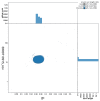
We present a genome assembly from a specimen of Danio rerio (zebra danio; Chordata; Actinopteri; Cypriniformes; Danionidae). The genome sequence has a total length of 1 413.66 megabases. Most of the assembly (99.85%) is scaffolded into 25 chromosomal pseudomolecules. The mitochondrial genome was also assembled, with a length of 16.6 kilobases. Gene annotation of this assembly on Ensembl identified 25 582 protein-coding genes.
Keywords: Cypriniformes; Danio rerio; chromosomal; genome sequence; zebra danio.
Copyright: © 2025 Howe K et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


References
-
- Challis R, Kumar S, Sotero-Caio C, et al. : Genomes on a Tree (GoaT): a versatile, scalable search engine for genomic and sequencing project metadata across the eukaryotic Tree of Life [version 1; peer review: 2 approved]. Wellcome Open Res. 2023;8:24. 10.12688/wellcomeopenres.18658.1 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

