This is a preprint.
Predicting Alzheimer's Disease Progression from Sparse Multimodal Data by NeuralODE Models
- PMID: 40909652
- PMCID: PMC12407814
- DOI: 10.1101/2025.08.26.672412
Predicting Alzheimer's Disease Progression from Sparse Multimodal Data by NeuralODE Models
Abstract
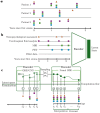
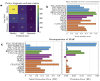
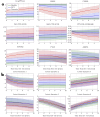
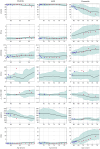
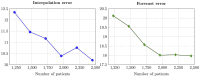
Alzheimer's disease shows significantly variable progressions between patients, making early diagnosis, disease monitoring, and care planning difficult. Existing data-driven Disease Progression Models try to tackle this issue, but they usually require sufficiently large datasets of specific diagnostic modalities, which are rarely available in clinical practice. Here, we introduce a new modeling framework capable of predicting individual disease trajectories from sparse, irregularly sampled, multi-modal clinical data. Our method uses (recurrent) Neural Ordinary Differential Equations to determine the current hidden state of a patient from sparse past exams and to forecast future disease progression, illustrating how biomarkers evolve over time. When applied to the ADNI clinical cohort, the model detected early signs of disease more accurately than common data-driven alternatives and effectively tracked changes in biomarker trajectories that align with established clinical knowledge. This provides a versatile tool for accurate diagnosis and monitoring of neurodegenerative diseases.
Keywords: Alzheimer’s disease; disease progression modeling; machine learning; neurodegenerative disorders; patient-specific trajectories; sparse and multimodal clinical data.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
