This is a preprint.
Catalytic pocket of Clr4 (Suv39h) methyltransferase serves as a substrate receptor for Cullin 4-dependent histone H3 ubiquitination
- PMID: 40909655
- PMCID: PMC12407891
- DOI: 10.1101/2025.08.28.672867
Catalytic pocket of Clr4 (Suv39h) methyltransferase serves as a substrate receptor for Cullin 4-dependent histone H3 ubiquitination
Abstract
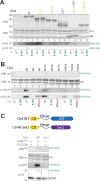
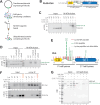
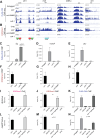
Histone H3 lysine 9 (H3K9) methylation must be regulated to prevent inappropriate heterochromatin formation. Regulation of the conserved fission yeast H3K9 methyltransferase Clr4 (Suv39h) involves an automethylation-induced conformational switch and interaction of its catalytic SET domain with mono-ubiquitinated histone H3 lysine 14 (H3K14ub), a modification catalyzed by the Cul4 subunit of the CLRC complex. Using reconstituted CLRC, we show that Clr4 catalytic pocket serves as a substrate receptor for Cul4-dependent H3K14 ubiquitination. H3K14ub activates Clr4 to catalyze cis methylation of H3K9 on the same histone tail, while Clr4 automethylation enables H3K14ub-bound Clr4 to methylate H3K9 on an unmodified H3 tail in trans. Crosslinking and structural modeling reveal interactions between Clr4 chromo and SET domains, and between the chromodomain and H3K14ub, suggesting that the chromodomain reads H3K9me3 and H3K14ub to allosterically regulate Clr4 activity. H3K14 ubiquitination therefore regulates Clr4 by promoting its recruitment and by positioning H3K9 in the active site.
Conflict of interest statement
Competing interest statement EpiCypher is a commercial developer and supplier of reagents (e.g., semisynthetic nucleosomes) used in this study. EFP, TW, MAC and M.-C.K. own shares in EpiCypher with M.-C.K. also a board member of same. All other authors declare no competing interests.
Figures




References
-
- Bühler M. & Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol 14, 1041–1048 (2007). - PubMed
-
- Jia S., Kobayashi R. & Grewal S. I. S. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol 7, 1007–1013 (2005). - PubMed
-
- Hong E. J. E., Villén J., Gerace E. L., Gygi S. P. & Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol 2, 106–111 (2005). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
