Identification and expression analysis of the BnE2F/DP gene family in Brassica napus
- PMID: 40909893
- PMCID: PMC12405181
- DOI: 10.3389/fpls.2025.1641897
Identification and expression analysis of the BnE2F/DP gene family in Brassica napus
Abstract
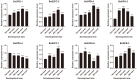
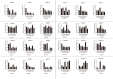
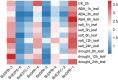
E2F/DP (Eukaryotic Transcription Factor 2/Dimerization Partner) refers to a class of protein complexes that play a pivotal role in the regulation of gene transcription in eukaryotes. In higher plants, E2F/DP transcription factors are of vital significance in mediating responses to environmental stresses. Based on differences in their conserved structural domains, they can be categorized into three subgroups: E2F, DP, and DEL (DP-E2F-like). Studies on E2F/DP in plants are relatively scarce, and no research on E2F/DP in Brassica napus has been reported to date. Utilize bioinformatics approaches to identify the BnE2F/DP gene family in Brassica napus. Construct a phylogenetic tree for evolutionary relationship analysis, and analyze the chromosomal distribution, gene structure composition, types of cis-acting elements, as well as intragenomic and interspecific collinearity. Integrate transcriptome data with real-time quantitative PCR (RT-qPCR) technology to explore the expression patterns of BnE2F/DP gene family members across various tissues. Total 29 BnE2F/DP genes were identified in Brassica napus and classified into 8 sub-families. These genes were unevenly distributed across 15 chromosomes, with enrichment on chromosomes 3 and 13. Intragenomic collinearity analysis detected 35 pairs of collinear BnE2F/DP gene pairs. Interspecific analysis revealed that there were 9 pairs of orthologous gene pairs between Brassica napus and Arabidopsis thaliana. Cis-acting element analysis showed that members of the BnE2F/DP family harbored four types of cis-acting elements, which might be involved in multiple regulatory processes, including hormonal regulation, abiotic stress responses, light reactions, and growth and development. Transcriptome and quantitative PCR data analyses indicate that members of the E2FC subfamily are likely to actively regulate seed and embryo development and positively respond to various abiotic stresses.. The conserved motifs and gene structures within each BnE2F/DP subfamily are largely consistent, which indicates that BnE2F/DP plays a crucial role in the growth and development of rapeseed seeds and embryos, as well as in the response to various abiotic stresses.
Keywords: Brassica napus; E2F/DP; bioinformatics; expression analysis; gene family.
Copyright © 2025 Shen, Wei and Fu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Berckmans B., Vassileva V., Schmid S. P., Maes S., Parizot B., Naramoto S., et al. (2011). Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 23, 3671–3683. doi: 10.1105/tpc.111.088377, PMID: - DOI - PMC - PubMed
-
- Biłas R., Szafran K., Hnatuszko-Konka K., Kononowicz A. K. (2016). Cis-regulatory elements used to control gene expression in plants. Plant Cell Tissue Organ Culture (PCTOC) 127, 269–287. doi: 10.1007/s11240-016-1057-7 - DOI
LinkOut - more resources
Full Text Sources

