Metabolomics reveal distinct molecular pathways associated with future risk of Crohn's Disease
- PMID: 40910526
- PMCID: PMC12416195
- DOI: 10.1080/19490976.2025.2546998
Metabolomics reveal distinct molecular pathways associated with future risk of Crohn's Disease
Abstract
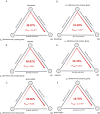
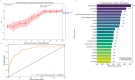
Host - microbiome interactions are central to Crohn'sdisease (CD) pathogenesis; yet the early metabolic alterations that precededisease onset remain poorly defined. To explore preclinical metabolicsignatures of CD, we analyzed baseline serum metabolomic profiles in a nestedcase-control study within the Crohn's and Colitis Canada - Genetics, Environment, Microbiome (CCC-GEM) Project, a prospective cohort of 5,122 healthyfirst-degree relatives (FDRs) of CD patients. We included 78 individuals wholater developed CD and 311 matched FDRs who remained disease-free. In an untargetedassessment of metabolomic data, we identified 63 metabolites significantlyassociated with future CD risk. Integrative analyses further identifiedmultiple associations between CD-related metabolites and proteomic markers, gutmicrobiome composition, antimicrobial antibody, fecal calprotectin andC-reactive protein. Quinolinate, a tryptophan catabolite, was elevated inindividuals who later developed CD and showed strong positive correlations withC-reactive protein, fecal calprotectin, and C-X-C motif chemokine ligand 9 (CXCL9).In contrast, higher levels of ascorbate and isocitrate were associated withreduced CD risk and were negatively correlated with C-reactive protein and CD-associated proteins.These findings identify several distinct molecular pathways that contribute toCD pathogenesis.
Keywords: Inflammatory bowel disease; Ruminococcus torques; gut barrier function; risk biomarkers.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390(10114):2769–18. doi: 10.1016/S0140-6736(17)32448-0. - DOI - PubMed
-
- Lee SH, Turpin W, Espin-Garcia O, Raygoza Garay JA, Smith MI, Leibovitzh H, Goethel A, Turner D, Mack D, Deslandres C, et al. Anti-microbial antibody response is associated with future onset of Crohn’s disease independent of biomarkers of altered gut barrier function, subclinical inflammation, and genetic risk. Gastroenterology. 2021;161(5):1540–1551. doi: 10.1053/j.gastro.2021.07.009. - DOI - PubMed
-
- Xue M, Leibovitzh H, Jingcheng S, Neustaeter A, Dong M, Xu W, Espin-Garcia O, Griffiths AM, Steinhart AH, Turner D, et al. Environmental factors associated with risk of Crohn’s disease development in the Crohn’s and Colitis Canada - genetic, environmental, microbial Project. Clin Gastroenterol Hepatol. 2024;22(9):1889–1897.e12. doi: 10.1016/j.cgh.2024.03.049. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
