Gene expression profiling and pathway analysis in acute myeloid leukaemia-normal karyotype patients
- PMID: 40911615
- PMCID: PMC12412999
- DOI: 10.1371/journal.pone.0328911
Gene expression profiling and pathway analysis in acute myeloid leukaemia-normal karyotype patients
Abstract
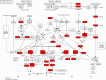
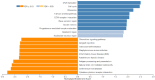
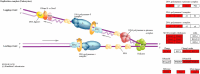
Acute myeloid leukaemia-normal karyotype (AML-NK) exhibits heterogeneity in expression profiles, influencing the treatment response and survival outcome. Transcriptome sequencing allows a comprehensive analysis of differentially expressed genes (DEGs) and dysregulated pathways in AML-NK, shedding light on the molecular mechanisms and their implications in patients' management. DEG analyses utilising transcriptome sequencing were conducted using a customised DESeq2 pipeline on 51 AML-NK patients at diagnosis (DX), 12 AML-NK patients who attained first remission (CR1) and 12 healthy controls. The transcriptomic sequencing of AML-NK compared to healthy controls revealed 5,126 DEGs, comprising 85.8% coding genes and 14.2% non-coding elements across 37 pathway categories. The AML-NK DX versus CR1 identified 5,621 DEGs consisting of 84.7% coding genes and 15.3% non-coding elements affecting 20 categories of pathways. Gene set enrichment analysis in this study revealed consistent upregulation of proliferative pathways, including cell cycle and DNA replication. In contrast, immune-related pathways, such as cytokine-cytokine receptor interactions and MHC antigen presentation pathways, were downregulated. Overexpression of oncogenes (FLT3, MYB, DNMT3B, and MYCN) in DX vs CR1 samples reinforces their usefulness in minimal residual disease monitoring, especially in AML-NK with no genetic aberrations. These findings reiterate the known hallmarks of cancers and validate the transcriptomic dysregulation in the pathogenesis of AML-NK. The robustness of the transcriptome sequencing findings was confirmed by RT-qPCR validation of six genes that were not reported in AML-NK patients. The comprehensive analyses of pathways with dysregulation of a myriad of genes led to an understanding of AML-NK pathogenesis and highlighted the markers for minimal residual disease. In summary, this study performed the first transcriptome-wide analysis of AML-NK in a Malaysian cohort and underscored pathways that are candidates for therapeutic interventions.
Copyright: © 2025 Ambayya et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
All authors declare that there is no competing interest.
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

