PTPN22-CD45 dual phosphatase retrograde feedback enhances TCR signaling and autoimmunity
- PMID: 40911684
- PMCID: PMC12412669
- DOI: 10.1126/sciadv.adw2568
PTPN22-CD45 dual phosphatase retrograde feedback enhances TCR signaling and autoimmunity
Abstract
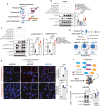
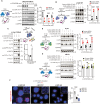
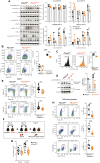
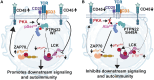
Protein tyrosine phosphatase nonreceptor type 22 (PTPN22) is encoded by a gene strongly associated with lupus and other autoimmune diseases. PTPN22 regulates T cell receptor (TCR) signaling through dephosphorylation of the kinases lymphocyte-specific protein tyrosine kinase (LCK) and zeta-chain-associated protein kinase 70 (ZAP70). The regulation of PTPN22 remains poorly understood. Here, we identify PTPN22 Ser449 as a protein kinase A phosphorylation site, which is triggered by TCR engagement and is hyperphosphorylated in lupus peripheral blood cells. PTPN22 Ser449 phosphorylation selectively lowered the affinity of PTPN22 for ZAP70 versus LCK but also indirectly suppressed inhibitory LCK Tyr192 phosphorylation through a ZAP70-CD45 signaling axis. The resulting dephosphorylation of LCK Tyr192 not only enhanced TCR signaling but also modulated pathway activation downstream the TCR. In vivo loss of PTPN22 Ser449 phosphorylation reduced T cell responses and suppressed experimental lupus nephritis. These results suggest that PTPN22 Ser449 phosphorylation promotes a CD45-mediated retrograde ZAP70-LCK feedback loop that enhances T cell responses and promotes autoimmunity.
Figures






References
-
- Moulton V. R., Lo M. S., Tsokos G. C., Methods and protocols to study T cell signaling abnormalities in human systemic lupus erythematosus. Methods Mol. Biol. 900, 25–60 (2012). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

