Phase separation of ERCC6L2-CtIP regulates the extent of DNA end resection
- PMID: 40913148
- PMCID: PMC12527916
- DOI: 10.1038/s41556-025-01760-4
Phase separation of ERCC6L2-CtIP regulates the extent of DNA end resection
Abstract
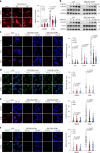
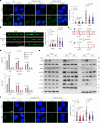
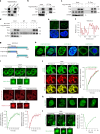
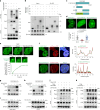
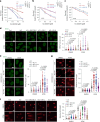
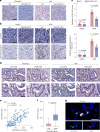
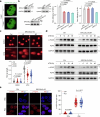
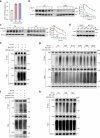
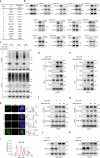
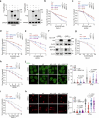
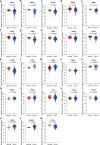
The ataxia telangiectasia mutated (ATM) kinase orchestrates the early stages of DNA double-strand break repair by promoting hyperphosphorylation of CtIP, a key step in the initiation of DNA end resection. However, the regulatory mechanisms controlling resection extent remain incompletely understood. Here we identify ERCC6L2 as a key regulator of DNA end resection in response to ATM inhibition. ERCC6L2 undergoes liquid-liquid phase separation via its intrinsically disordered regions, forming dynamic nuclear condensates that regulate CtIP stability. Disruption of these condensates renders CtIP susceptible to RNF138-mediated ubiquitination and degradation, thereby mitigating the heightened chromatin recruitment of CtIP induced by ATM inhibition. Intriguingly, ERCC6L2 is frequently downregulated in multiple cancer types and correlates with resistance to ATM inhibitors in both in vitro and in vivo settings. Our findings unveil the crucial role of ERCC6L2-CtIP condensates in governing the extent of DNA end resection and underscore the potential significance of ERCC6L2 as a predictive biomarker for ATM inhibitor response.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

