Feasibility and clinical utility of expanded genomic newborn screening in the Early Check program
- PMID: 40913169
- PMCID: PMC12520013
- DOI: 10.1038/s41591-025-03945-8
Feasibility and clinical utility of expanded genomic newborn screening in the Early Check program
Abstract
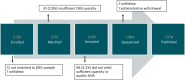
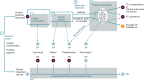
Although genomic sequencing presents groundbreaking newborn screening (NBS) opportunities, critical feasibility and utility questions remain. Here we present initial results from the Early Check program-an observational study assessing the feasibility and clinical utility of genomic NBS in North Carolina. Recruitment was statewide through mailed letters with electronic consent. Genome sequencing with analysis of 169 high actionability genes (plus 29 optional lower actionability genes) was performed using residual NBS dried blood spots. In 8 months, 1,979 newborns were screened, with 50 (2.5%) screen positives. Negative results were returned electronically, positive results by genetic counselors. Twenty-eight results (55%) were true positives, all received anticipatory guidance, surveillance and management recommendations, and referral to specialists as appropriate. We report technical feasibility and preliminary clinical utility finding, along with interpretation and follow-up challenges that hinder public health implementation. We propose standardized terminology to facilitate cross-study comparisons and accurate characterization of genomic NBS outcomes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: R.S.Z., S.F.S., A.B., A.J.G., K.G.L., K.G.M., K.M., C.K., K.S.H. and P.K. were employees of GeneDx, LLC at the time of this study and may have held company stock. All RTI authors (H.L.C., K.S.K., H.E.F., A.Y.G., A.N.F., B.A.M., B.W., R.R.M., D.B.B.J., A.C.W., M.R. and H.L.P.) were supported in this research by Janssen Pharmaceuticals (July 2021 to July 2022), Breakthrough T1D (formerly JDRF International), the Leona M. and Harry B. Helmsley Charitable Trust, Travere Therapeutics and Orchard Therapeutics. UNC authors (E.R.J., J.A.S. and C.M.P.) were supported in this research by Breakthrough T1D (formerly JDRF International). The other authors declare no competing interests.
Figures
References
MeSH terms
Grants and funding
- UL1 TR002489/TR/NCATS NIH HHS/United States
- UM1 TR004406/TR/NCATS NIH HHS/United States
- UM1TR004406/U.S. Department of Health & Human Services | NIH | National Center for Advancing Translational Sciences (NCATS)
- UL1TR002489/U.S. Department of Health & Human Services | NIH | National Center for Advancing Translational Sciences (NCATS)
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

