Lung virome convergence precedes hospital-acquired pneumonia in intubated critically ill patients
- PMID: 40914165
- PMCID: PMC12490257
- DOI: 10.1016/j.xcrm.2025.102289
Lung virome convergence precedes hospital-acquired pneumonia in intubated critically ill patients
Abstract
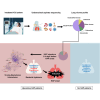
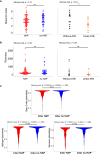
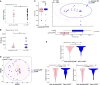
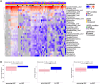
Hospital-acquired pneumonia (HAP) is one of the most common nosocomial infections, leading to significant morbidity and mortality in critically ill patients. HAP is previously associated with dysbiosis of the microbiota. However, the composition of the lung virome and its role in HAP pathogenesis remain unclear. Here, we longitudinally analyze the endotracheal virome in 87 critically ill patients, including 48 with HAP. Within the virome dominated by Caudoviricetes, a decrease in viral beta-diversity toward a bacteriophage-dominated signature and a distinct viral-bacterial interactome is observed 5-4 days before HAP onset. Lung virome composition, viral convergence before HAP onset, and conservation of 18% of the bacteriophage signature are validated in an external cohort of 40 patients. In silico causal inference further identifies bacteriophages associated with Streptococcus and Prevotella as a key regulator of HAP onset. These findings suggest an uncovered pathophysiological mechanism of HAP with virome involvement in lung microbiota dysbiosis. The discovery and validation studies are registered at ClinicalTrials.gov (NCT02003196 and NCT04793568).
Keywords: Caudoviricetes bacteriophages; Respiratory virome; cross-kingdom interactions; hospital-acquired pneumonia (HAP); intubated critically ill patients; microbiome; respiratory interactome; viral metagenomics.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- He Q., Wang W., Zhu S., Wang M., Kang Y., Zhang R., Zou K., Zong Z., Sun X. The epidemiology and clinical outcomes of ventilator-associated events among 20,769 mechanically ventilated patients at intensive care units: an observational study. Crit. Care. 2021;25:44. doi: 10.1186/s13054-021-03484-x. - DOI - PMC - PubMed
-
- Montassier E., Kitsios G.D., Radder J.E., Le Bastard Q., Kelly B.J., Panzer A., Lynch S.V., Calfee C.S., Dickson R.P., Roquilly A. Robust airway microbiome signatures in acute respiratory failure and hospital-acquired pneumonia. Nat. Med. 2023;29:2793–2804. doi: 10.1038/s41591-023-02617-9. - DOI - PubMed
-
- Bal A., Pichon M., Picard C., Casalegno J.S., Valette M., Schuffenecker I., Billard L., Vallet S., Vilchez G., Cheynet V., et al. Quality control implementation for universal characterization of DNA and RNA viruses in clinical respiratory samples using single metagenomic next-generation sequencing workflow. BMC Infect. Dis. 2018;18:537. doi: 10.1186/s12879-018-3446-5. - DOI - PMC - PubMed
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical

