ctDNA detects residual disease after neoadjuvant chemoradiotherapy and guides adjuvant therapy in esophageal squamous cell carcinoma
- PMID: 40914168
- PMCID: PMC12490249
- DOI: 10.1016/j.xcrm.2025.102334
ctDNA detects residual disease after neoadjuvant chemoradiotherapy and guides adjuvant therapy in esophageal squamous cell carcinoma
Abstract
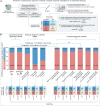
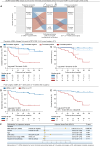
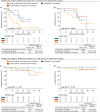
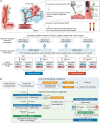
The diagnostic accuracy of circulating tumor DNA (ctDNA) for detecting molecular residual disease (MRD) after multimodal treatment remains unclear. In a prospective cohort of 132 patients with locally advanced esophageal squamous cell carcinoma (ESCC) undergoing neoadjuvant chemoradiotherapy (nCRT) followed by clinical response evaluation and surgery, tumor-informed personalized-panel and fixed-panel ctDNA assays are applied to serial blood samples. Personalized ctDNA assay demonstrates a superior baseline detection rate (99.2%) and outperforms fixed panels in diagnosing post-nCRT residual disease. Integrating personalized ctDNA with conventional clinical diagnostic methods increases sensitivity for predicting non-pathological complete response (non-pCR) from 78.4%-80.7% to 92.0%-93.2%. Patients with detectable MRD post-nCRT and/or post-surgery exhibit worse survival outcomes. In non-pCR patients, adjuvant immunotherapy improves disease-free survival in post-surgery MRD-positive cases, whereas MRD-negative patients derive no benefit. These findings support incorporating ctDNA into response assessment to guide organ-sparing strategies and adjuvant therapy decisions in ESCC. This study is registered at ClinicalTrials.gov (NCT03937362).
Keywords: adjuvant immunotherapy; circulating tumor DNA; esophageal squamous cell carcinoma; molecular residual disease; neoadjuvant chemoradiotherapy; organ preservation.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests X.G. has received a personal research grant from the Nijbakker-Morra Foundation. G.W., P.C., X.F., J.Y., Z.Z., and S.C. are employees of Burning Rock Biotech.
Figures



References
-
- van Hagen P., Hulshof M.C.C.M., van Lanschot J.J.B., Steyerberg E.W., van Berge Henegouwen M.I., Wijnhoven B.P.L., Richel D.J., Nieuwenhuijzen G.A.P., Hospers G.A.P., Bonenkamp J.J., et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N. Engl. J. Med. 2012;366:2074–2084. doi: 10.1056/NEJMoa1112088. - DOI - PubMed
-
- Yang H., Liu H., Chen Y., Zhu C., Fang W., Yu Z., Mao W., Xiang J., Han Y., Chen Z., et al. Neoadjuvant Chemoradiotherapy Followed by Surgery Versus Surgery Alone for Locally Advanced Squamous Cell Carcinoma of the Esophagus (NEOCRTEC5010): A Phase III Multicenter, Randomized, Open-Label Clinical Trial. J. Clin. Oncol. 2018;36:2796–2803. doi: 10.1200/JCO.2018.79.1483. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical

