Predominance of L. monocytogenes Lineage I Clones in Wastewater, Ruminants, and Natural Environments
- PMID: 40916165
- PMCID: PMC12415317
- DOI: 10.1111/1462-2920.70169
Predominance of L. monocytogenes Lineage I Clones in Wastewater, Ruminants, and Natural Environments
Abstract
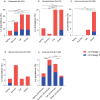
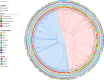
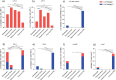
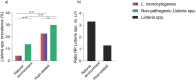
Listeria monocytogenes is a saprophytic bacterium and a foodborne pathogen of humans and animals. Little is known about its distribution and genetic diversity across different environments within the same geographical region. We conducted a large-scale longitudinal study in southeastern Spain monitoring Listeria spp. in untreated wastewater, ruminant farms, and natural environments over four seasons (N = 1490 samples, N = 545 isolates) and in food and food-processing environments (N = 7395 samples, N = 255 isolates). Listeria spp. were more abundant in host-associated than natural environments, and non-pathogenic Listeria were more prevalent than L. monocytogenes in both niches. L. monocytogenes was detected in 42.7%, 11.4%, 4.2%, and 3.4% of wastewater, ruminant farms, natural environments, and food-related samples, respectively. Hypervirulent lineage I accounted for 82.9% of L. monocytogenes isolates from wastewater, ruminant farms, and natural environments, while lineage II represented 74.1% in food-related samples. Among 255 L. monocytogenes cgMLST types, 5% were shared across environments, demonstrating circulation between different environments. Persistent L. monocytogenes clones were detected in food processing environments and ruminant farms. Our data suggest anthropogenic activities and livestock drive Listeria spp. dissemination. These results provide insights into the interactions of Listeria spp. in the environment, improving surveillance strategies to reduce pathogen transmission, food contamination, and clinical cases.
Keywords: cgMLST; ecology; foodborne; pathogens; persistence; virulence; whole genome sequencing.
© 2025 The Author(s). Environmental Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have nothing to report.
The authors declare no conflicts of interest.
Figures


References
-
- Bagatella, S. , Tavares‐Gomes L., and Oevermann A.. 2022. “ <styled-content style="fixed-case"> Listeria monocytogenes </styled-content> at the Interface Between Ruminants and Humans: A Comparative Pathology and Pathogenesis Review.” Veterinary Pathology 59: 186–210. 10.1177/03009858211052659. - DOI - PubMed
-
- Castro, H. , Jaakkonen A., Hakkinen M., Korkeala H., and Lindström M.. 2018. “Occurrence, Persistence, and Contamination Routes of <styled-content style="fixed-case"> Listeria monocytogenes </styled-content> Genotypes on Three Finnish Dairy Cattle Farms: A Longitudinal Study.” Applied and Environmental Microbiology 84: 84. 10.1128/AEM.02000-17. - DOI - PMC - PubMed
-
- Chapin, T. K. , Nightingale K. K., Worobo R. W., Wiedmann M., and Strawn L. K.. 2014. “Geographical and Meteorological Factors Associated With Isolation of Listeria Species in New York State Produce Production and Natural Environments.” Journal of Food Protection 77: 1919–1928. 10.4315/0362-028X.JFP-14-132. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- CIAICO/2023/053/Generalitat Valenciana
- PID2022-137961OB-I00/Ministerio de Ciencia, Innovación y Universidades
- RYC-2018-024985-I/Ministerio de Ciencia, Innovación y Universidades
- PID2023-152404OB-I00/Ministerio de Ciencia, Innovación y Universidades
- RYC2021-032245-I/Ministerio de Ciencia, Innovación y Universidades
LinkOut - more resources
Full Text Sources
Miscellaneous

