Role of CPEBs in Learning and Memory
- PMID: 40916649
- PMCID: PMC12415542
- DOI: 10.1111/jnc.70226
Role of CPEBs in Learning and Memory
Abstract
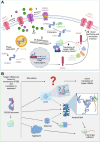
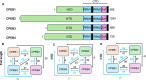
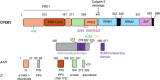
Memory formation involves a complex interplay of molecular and cellular processes, including synaptic plasticity mechanisms such as long-term potentiation (LTP) and long-term depression (LTD). These processes rely on activity-dependent gene expression and local protein synthesis at synapses. A central unresolved question in neuroscience is how memories can be stably maintained over time, despite the transient nature of the proteins involved in their initial encoding. A key candidate addressing this 'maintenance paradox' is the CPEB (cytoplasmic polyadenylation element-binding protein) family, particularly CPEB3. CPEBs are RNA-binding proteins that regulate the polyadenylation and translation of dormant mRNAs, enabling synaptic tagging and memory consolidation. CPEB3 has been shown to modulate the expression of critical synaptic proteins, including AMPA and NMDA receptor subunits, thereby influencing synaptic strength and long-term memory persistence. Structurally, CPEB3 features a disordered N-terminal domain (NTD) enriched in glutamine and proline residues, which may facilitate reversible aggregation and phase separation and an actin-binding domain, potentially supporting its localisation to ribonucleoprotein granules. The highly conserved C-terminal domain (CTD) contains RNA-recognition motifs essential for mRNA binding. Together, these structural features may enable CPEB3 to function as a molecular switch, linking synaptic activity to enduring changes in protein synthesis and memory encoding. Here, we review the current understanding of the function of CPEB3, highlighting current hypotheses and debates of the role(s) of protein self-assembly in memory formation.
Keywords: AMPA; CPEB; RNA‐binding protein; amyloid; memory; neuroscience.
© 2025 The Author(s). Journal of Neurochemistry published by John Wiley & Sons Ltd on behalf of International Society for Neurochemistry.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Atkins, C. M. , Davare M. A., Oh M. C., Derkach V., and Soderling T. R.. 2005. “Bidirectional Regulation of Cytoplasmic Polyadenylation Element‐Binding Protein Phosphorylation by Ca2+/Calmodulin‐Dependent Protein Kinase II and Protein Phosphatase 1 During Hippocampal Long‐Term Potentiation.” Journal of Neuroscience 25, no. 23: 5604–5610. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

