Population Genomics Reveals Panmixia in Pacific Sardine (Sardinops sagax) of the North Pacific
- PMID: 40917341
- PMCID: PMC12409727
- DOI: 10.1111/eva.70154
Population Genomics Reveals Panmixia in Pacific Sardine (Sardinops sagax) of the North Pacific
Abstract
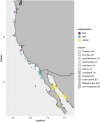
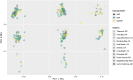
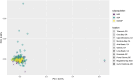
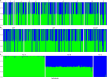
The spatial structure and dynamics of populations are important considerations when defining management units in organisms that are harvested as natural resources. In the Eastern Pacific, Pacific Sardine range from Chile to Alaska, the northernmost state of the United States (U.S.), and once supported an expansive and productive fishery. Along its North American range, it is hypothesized to comprise three subpopulations: a northern and southern subpopulation, which primarily occur off the coast of the U.S. and Baja California, Mexico (M.X.), respectively, and a third in the Gulf of California, M.X. We used low coverage whole genome sequencing to generate genotype likelihoods for millions of SNPs in 317 individuals collected from the Gulf of California, M.X., to Oregon, U.S., to assess population structure in Pacific Sardine. Differentiation across the genome was driven by variation at several putative chromosomal inversions ranging in size from ~21 MB to 0.89 MB, although none of the putative inversions showed any evidence of geographic differentiation. Our results support panmixia across an impressive ~4000 km range.
Keywords: Alosidae; California current; Clupeiformes; fisheries genomics; population structure.
© 2025 The Author(s). Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Adams, E. S. , and Craig M. T.. 2024. “Phylogeography of the Pacific Sardine, Sardinops sagax, Across Its Northeastern Pacific Range.” Bulletin of the Southern California Academy of Sciences 123: 10–24. 10.3160/0038-3872-123.1.10. - DOI
-
- Ahlstrom, E. H. 1954. Distribution and Abundance of Egg and Larval Populations of the Pacific Sardine (Fishery Bulletin 93 No. Volume 56). From Fishery Bulletin of the Fish and Wildlife Service.
-
- Andrews, S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data.
LinkOut - more resources
Full Text Sources
Miscellaneous

