Advances in solid-state NMR methods for studying RNA structures and dynamics
- PMID: 40918042
- PMCID: PMC12406530
- DOI: 10.1016/j.mrl.2024.200133
Advances in solid-state NMR methods for studying RNA structures and dynamics
Abstract
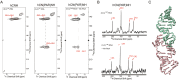
Ribonucleic acid (RNA) structures and dynamics play a crucial role in elucidating RNA functions and facilitating the design of drugs targeting RNA and RNA-protein complexes. However, obtaining RNA structures using conventional biophysical techniques, such as X-ray crystallography and solution nuclear magnetic resonance (NMR), presents challenges due to the inherent flexibility and susceptibility to degradation of RNA. In recent years, solid-state NMR (SSNMR) has rapidly emerged as a promising alternative technique for characterizing RNA structure and dynamics. SSNMR has several distinct advantages, including flexibility in sample states, the ability to capture dynamic features of RNA in solid form, and suitability to character RNAs in various sizes. Recent decade witnessed the growth of 1H-detected SSNMR methods on RNA, which targeted elucidating RNA topology and base pair dynamics in solid state. They have been applied to determine the topology of RNA segment in human immunodeficiency virus (HIV) genome and the base pair dynamics of riboswitch RNA. These advancements have expanded the utility of SSNMR techniques within the RNA research field. This review provides a comprehensive discussion of recent progress in 1H-detected SSNMR investigations into RNA structure and dynamics. We focus on the established 1H-detected SSNMR methods, sample preparation protocols, and the implementation of rapid data acquisition approaches.
Keywords: Dynamics; Pulse sequences; RNA; Solid-state NMR; Structure.
© 2024 The Authors.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Shenlin Wang is an editorial board member of Magnetic Resonance Letters but was not involved in the editorial review or the decision to publish this article.
Figures







References
-
- Selmer M., Dunham C.M., Murphy F.V., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. - PubMed
-
- Anger A.M., Armache J.-P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. - PubMed
-
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., le Noble F., Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
