Expressed mutated genes in Sezary syndrome and their potential prognostic value in patients treated with extracorporeal photopheresis
- PMID: 40918137
- PMCID: PMC12411188
- DOI: 10.3389/fimmu.2025.1589467
Expressed mutated genes in Sezary syndrome and their potential prognostic value in patients treated with extracorporeal photopheresis
Abstract
Background: Sézary syndrome (SS) is an aggressive and leukemic variant of Cutaneous T-cell Lymphoma (CTCL) with an incidence of 1 case per million people per year. It is characterized by a complex and heterogeneous profile of genetic alteration ns that has so far precluded the development of a specific and definitive therapeutic intervention.
Methods: Deep-RNA-sequencing (RNA-seq) data were used to analyze the single nucleotide variants (SNVs) carried by 128 putative CTCL-driver genes, previously identified as mutated in genomic studies, in longitudinal SS samples collected from 17 patients subjected to extracorporeal photopheresis (ECP) with Interferon-α. Results obtained were integrated with Whole Exome Sequencing (WES) data. SNVs were validated using the Sanger method. Pathway analysis was performed with g:Profiler web server (https://biit.cs.ut.ee/gprofiler/gost). Statistical analyses were performed with GraphPad PRISM 8 software.
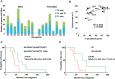
Results: Nonsynonymous SNVs were identified in 56 genes. Integration of RNA-seq with WES data revealed that about half of these genes contained somatic mutations. Among them, the most frequently transcribed mutated genes were TET2, JAK3, NCOR1, PDCD11, RHOA, and TP53. Nearly all the remaining genes had germline-restricted mutations, and included ARID1A, ATM, ATR, CREBBP, POLD1, and POT1 genes, which are involved in DNA repair, homologous recombination, and chromatin remodeling, and the CROCC gene, implicated in centrosome cohesion. Monitoring of the mutated genes, identified within an enlarged panel of CTCL associated genes, revealed their reduction in almost 70% of SS patients as well as a significant decline of total number of mutations (SNVs) during ECP treatment. Several mutated genes persisted post-therapy, representing novel candidates associated with ECP resistance that could also have a potential prognostic relevance. Notably, these genes mainly converge on pathways related to DNA repair (ATR, ATRIP, POLD1, TP53, TP53BP1/2) which might represent novel targets to be explored in combination with ECP.
Conclusions: This is the first evaluation in SS of expressed mutations in a large panel of CTCL-driver genes. Also innovative is the monitoring of mutated genes in patients' malignant lymphocytes during ECP, a first-line treatment of CTCL, which highlights novel candidates associated with ECP resistance that might unmask novel pharmacological vulnerabilities to be exploited during ECP for a personalized treatment.
Keywords: RNA-seq; Sezary syndrome; candidates associated with therapy resistance and personalized treatment; cutaneous T-cell lymphoma; extracorporeal photopheresis; whole exome sequencing.
Copyright © 2025 Cristofoletti, Salvatore, Bassi, Negrini, Scaglione, Mazzarella, Frigè, Minafò, Fioretti, Monopoli, Accetturi, Pilla, Di Raimondo, Frezzolini, Scala, D’Atri, Russo and Narducci.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Scarisbrick JJ, Prince HM, Vermeer MH, Quaglino P, Horwitz S, Porcu P, et al. Cutaneous lymphoma international consortium study of outcome in advanced stages of mycosis fungoides and sézary syndrome: effect of specific prognostic markers on survival and development of a prognostic model. J Clin Oncol. (2015) 33:3766–73. doi: 10.1200/JCO.2015.61.7142, PMID: - DOI - PMC - PubMed
-
- Caprini E, Cristofoletti C, Arcelli D, Fadda P, Citterich MH, Sampogna F, et al. Identification of key regions and genes important in the pathogenesis of Sézary syndrome by combining genomic and expression microarrays. Cancer Res. (2009) 69:8438–46. doi: 10.1158/0008-5472.CAN-09-2367, PMID: - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

