Cytotoxic CX3CR1+ Vδ1 T cells clonally expand in an interplay of CMV, microbiota, and HIV-1 persistence in people on antiretroviral therapy
- PMID: 40920867
- PMCID: PMC12431655
- DOI: 10.1371/journal.ppat.1013489
Cytotoxic CX3CR1+ Vδ1 T cells clonally expand in an interplay of CMV, microbiota, and HIV-1 persistence in people on antiretroviral therapy
Abstract
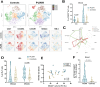
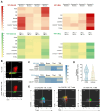
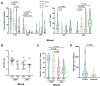
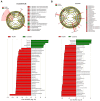
Vδ1 γδ T cells are key players in innate and adaptive immunity, particularly at mucosal interfaces such as the gut. An increase in circulating Vδ1 cells has long been observed in people with HIV-1, but remains poorly understood. We performed a comprehensive characterization of Vδ1 T cells in blood and duodenal intra-epithelial lymphocytes, obtained from endoscopic mucosal biopsies of 15 people with HIV-1 on antiretroviral therapy and 15 HIV-seronegative controls, in a substudy of the ANRS EP61 GALT study (NCT02906137). We deciphered the phenotype, functional profile, single-cell transcriptome and repertoire of Vδ1 cells and unraveled their relationships with the possible triggers involved, in particular CMV and microbiota. We also assessed whether Vδ1 T cells may play a role in controlling the HIV-1 reservoir. Vδ1 T cells were mainly terminally differentiated effectors that clonally expanded in the blood with some trafficking with the gut of people with HIV-1. Most expressed CX3CR1 and displayed a highly cytotoxic profile, but low cytokine production, supported by a transcriptomic shift towards enhanced effector lymphocytes. This expansion was associated with CMV status and markers of occult replication, but also with changes in the duodenal and blood-translocated microbiota. Cytotoxic, but not IFN-γ-producing, Vδ1 T cells were negatively associated with cell-associated HIV-1 RNA in both the blood and duodenal compartments. The increase in Vδ1 T cells observed in people with HIV-1 has multiple triggers, particularly CMV and microbiota, and may in turn contribute to the control of the HIV-1 reservoir.
Copyright: © 2025 Collercandy et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

