The DNA replication machinery transmits dual signals to prevent unscheduled licensing and execution of centrosome duplication
- PMID: 40921755
- PMCID: PMC12417547
- DOI: 10.1038/s41467-025-63002-3
The DNA replication machinery transmits dual signals to prevent unscheduled licensing and execution of centrosome duplication
Abstract
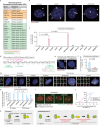
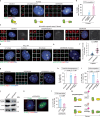
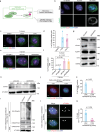
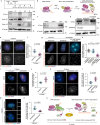
Copy number control of DNA and centrosomes is essential for accurate genetic inheritance. DNA replication and centrosome duplication have been recognized as parallel key events for cell division. Here, we discover that the DNA replication machinery directly regulates the licensing and execution processes of centrosome duplication to prevent centrosome amplification. We find that the microcephaly protein DONSON couples DNA replication initiation with Cdc6 translocation to centrosomes. The Cdc6 signal prevents the precocious occurrence of centriole disengagement, the licensing step for centrosome duplication. During DNA replication, DONSON inhibits replisome disassembly by interacting with the CMG helicase, maintaining the intrinsic S/G2 checkpoint signal that blocks centriole-to-centrosome conversion, the execution step for centrosome duplication. Disruption of these dual signals causes precocious centrosome duplication and chromosome mis-segregation, observed in DONSON patient cells. Our results reveal that the DNA replication machinery not only duplicates genetic material but also controls the system for its accurate segregation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

