Bayesian estimation of HIV acquisition dates for prevention trials
- PMID: 40923785
- PMCID: PMC12505913
- DOI: 10.1128/mbio.01881-25
Bayesian estimation of HIV acquisition dates for prevention trials
Abstract
Accurate timing estimates of when participants acquire HIV in HIV prevention trials are necessary for determining antibody levels at acquisition. The Antibody-Mediated Prevention (AMP) Studies showed that a passively administered broadly neutralizing antibody can prevent the acquisition of HIV from a neutralization-sensitive virus. We developed a pipeline for estimating the date of detectable HIV acquisition (DDA) in AMP Study participants using diagnostic and viral sequence data. Using a Bayesian strategy that combines three streams of data (REN [rev/vpu/env/Δnef] sequence, GP [gag/Δpol] sequence, and diagnostic) where their 95% credible intervals overlap based on pre-specified criteria and decision rules. We evaluated the performance of our AMP pipeline using PacBio viral sequence data from 41 participants across two prospective acute HIV acquisition cohort studies, FRESH and RV217, with twice-weekly sampling. These cohort studies enrolled young women in South Africa and men and women in Kenya and Thailand, respectively, with a high likelihood of HIV acquisition. In evaluating performance, "true DDA" was the center of bounds between last-negative and first-positive RNA diagnostic tests (median time 4 days, range 2-7 days); bias was the mean difference between estimated and true DDA. Using diagnostic data alone yielded timing estimates with a bias of 2.4 days and root mean square error (RMSE) of 7.9 days. These results were improved using sequence + diagnostic data (bias 1.5 days, RMSE 6.9 days), as well as by restricting sequence-based estimation to samples from ≤5 weeks post-DDA (bias 0.2 days, RMSE 7.8 days).IMPORTANCEIn HIV prevention trials, accurate timing estimates of when individual participants acquire HIV can be used to estimate antibody levels at the time of acquisition, which is useful for projecting antibody levels needed for prevention. The results we report here suggest that if sequence-based estimation of acquisition timing is used in future clinical trials of combination broadly neutralizing antibody (bnAb) regimens or multispecific bnAbs for HIV prevention, a sampling frequency of at least monthly is needed. Moreover, in the samples analyzed here, we observed less bias in sequence-based timing estimation for samples taken <5 weeks post-DDA. This observation is consistent with the timing of immune-driven selective pressures that may negatively impact the power to detect acquisition sieve effects.
Keywords: Antibody-Mediated Prevention (AMP) Studies; Bayesian posterior distribution; FRESH; HIV; RV217; acute acquisition cohort; date of detectable acquisition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
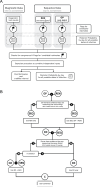

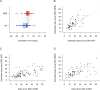
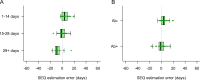
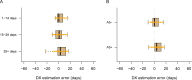



References
-
- Huang Y, Naidoo L, Zhang L, Carpp LN, Rudnicki E, Randhawa A, Gonzales P, McDermott A, Ledgerwood J, Lorenzo MMG, Burns D, DeCamp A, Juraska M, Mascola J, Edupuganti S, Mgodi N, Cohen M, Corey L, Andrew P, Karuna S, Gilbert PB, Mngadi K, Lazarus E. 2021. Pharmacokinetics and predicted neutralisation coverage of VRC01 in HIV-uninfected participants of the Antibody Mediated Prevention (AMP) trials. EBioMedicine 64:103203. doi: 10.1016/j.ebiom.2020.103203 - DOI - PMC - PubMed
-
- Huang Y, Zhang L, Eaton A, Mkhize NN, Carpp LN, Rudnicki E, DeCamp A, Juraska M, Randhawa A, McDermott A, Ledgerwood J, Andrew P, Karuna S, Edupuganti S, Mgodi N, Cohen M, Corey L, Mascola J, Gilbert PB, Morris L, Montefiori DC. 2022. Prediction of serum HIV-1 neutralization titers of VRC01 in HIV-uninfected antibody mediated prevention (AMP) trial participants. Hum Vaccin Immunother 18:1908030. doi: 10.1080/21645515.2021.1908030 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
