A multiomics recovery factor predicts long COVID in the IMPACC study
- PMID: 40924481
- PMCID: PMC12582403
- DOI: 10.1172/JCI193698
A multiomics recovery factor predicts long COVID in the IMPACC study
Abstract
BACKGROUNDFollowing SARS-CoV-2 infection, approximately 10%-35% of patients with COVID-19 experience long COVID (LC), in which debilitating symptoms persist for at least 3 months. Elucidating the biologic underpinnings of LC could identify therapeutic opportunities.METHODSWe utilized machine learning methods on biologic analytes provided over 12 months after hospital discharge from more than 500 patients with COVID-19 in the IMPACC cohort to identify a multiomics "recovery factor," trained on patient-reported physical function survey scores. Immune profiling data included PBMC transcriptomics, serum O-link and plasma proteomics, plasma metabolomics, and blood mass cytometry by time of flight (CyTOF) protein levels. Recovery factor scores were tested for association with LC, disease severity, clinical parameters, and immune subset frequencies. Enrichment analyses identified biologic pathways associated with recovery factor scores.RESULTSParticipants with LC had lower recovery factor scores compared with recovered participants. Recovery factor scores predicted LC as early as hospital admission, irrespective of acute COVID-19 severity. Biologic characterization revealed increased inflammatory mediators, elevated signatures of heme metabolism, and decreased androgenic steroids as predictive and ongoing biomarkers of LC. Lower recovery factor scores were associated with reduced lymphocyte and increased myeloid cell frequencies. The observed signatures are consistent with persistent inflammation driving anemia and stress erythropoiesis as major biologic underpinnings of LC.CONCLUSIONThe multiomics recovery factor identifies patients at risk of LC early after SARS-CoV-2 infection and reveals LC biomarkers and potential treatment targets.TRIAL REGISTRATIONClinicalTrials.gov NCT04378777.FUNDINGNational Institute of Allergy and Infectious Diseases (NIAID), NIH (3U01AI167892-03S2, 3U01AI167892-01S2, 5R01AI135803-03, 5U19AI118608-04, 5U19AI128910-04, 4U19AI090023-11, 4U19AI118610-06, R01AI145835-01A1S1, 5U19AI062629-17, 5U19AI057229-17, 5U19AI057229-18, 5U19AI125357-05, 5U19AI128913-03, 3U19AI077439-13, 5U54AI142766-03, 5R01AI104870-07S1, 3U19AI089992-09, 3U19AI128913-03, and 5T32DA018926-1, 3U19AI1289130, U19AI128913-04S1, R01AI122220); NIH (UM1TR004528); and National Science Foundation (NSF) (DMS2310836).
Keywords: Biomarkers; COVID-19; Immunology; Infectious disease; Machine learning.
Figures
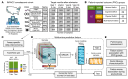


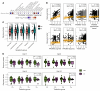


Update of
-
Identification of a multi-omics factor predictive of long COVID in the IMPACC study.bioRxiv [Preprint]. 2025 Feb 14:2025.02.12.637926. doi: 10.1101/2025.02.12.637926. bioRxiv. 2025. Update in: J Clin Invest. 2025 Sep 9;135(21):e193698. doi: 10.1172/JCI193698. PMID: 39990442 Free PMC article. Updated. Preprint.
References
-
- National Academies of Sciences E and Medicine. A Long COVID Definition: A Chronic, Systemic Disease State with Profound Consequences. The National Academies Press; 2024. - PubMed
MeSH terms
Associated data
Grants and funding
- U19 AI090023/AI/NIAID NIH HHS/United States
- U19 AI118608/AI/NIAID NIH HHS/United States
- U01 AI167892/AI/NIAID NIH HHS/United States
- U54 AI142766/AI/NIAID NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- U19 AI062629/AI/NIAID NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- U19 AI128910/AI/NIAID NIH HHS/United States
- R01 AI104870/AI/NIAID NIH HHS/United States
- T32 DA018926/DA/NIDA NIH HHS/United States
- U19 AI125357/AI/NIAID NIH HHS/United States
- R01 AI145835/AI/NIAID NIH HHS/United States
- U19 AI128913/AI/NIAID NIH HHS/United States
- R01 AI132774/AI/NIAID NIH HHS/United States
- R01 AI122220/AI/NIAID NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- R01 AI135803/AI/NIAID NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
- UM1 TR004528/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

