Virologic characteristics of SARS-CoV-2 infection across evolving Omicron subvariants
- PMID: 40924669
- PMCID: PMC12581670
- DOI: 10.1172/jci.insight.192228
Virologic characteristics of SARS-CoV-2 infection across evolving Omicron subvariants
Abstract
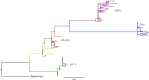
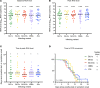
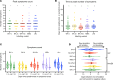
BACKGROUNDSARS-CoV-2 has evolved subvariants since the emergence of the Omicron variant in 2021. Whether these changes impact viral shedding and transmissibility is not known.METHODSPOSITIVES is a prospective longitudinal cohort of individuals with mild SARS-CoV-2 infection. Ambulatory, immunocompetent participants who did not receive antivirals self-administered 6 anterior nasal swabs over 15 days. Samples were analyzed by qPCR to quantify viral RNA, semiquantitative viral culture to detect shedding of replication-competent virus, and whole-genome sequencing to classify subvariants. Our predictor of interest was Omicron subvariants: BA.1x, BA.2x, BA.4/5x, XBB.x, and JN.x. Outcomes included RNA levels and duration of shedding replication-competent virus. We additionally explored whether symptoms are a valid marker for ending isolation.RESULTSThe median peak nasal SARS-CoV-2 RNA (6.0-6.3 log10 RNA copies/mL), median days to peak RNA (4-5 days), median days to undetectable viral RNA (12-14 days), and median days to negative viral culture (4-8 days) were similar across Omicron subvariants. Number and duration of symptoms were also similar. For all subvariants, a sizeable percentage (range 27.5%-56.0%) shed replication-competent virus after fever resolution and improvement of symptoms.CONCLUSIONDespite ongoing viral evolution, key aspects of viral dynamics of SARS-CoV-2 infection, including the duration of shedding replication-competent virus, have not substantially changed across Omicron subvariants. Replication-competent shedding of these subvariants is detected for a large proportion of people who meet criteria for ending isolation.FUNDINGNIH (U19 AI110818, R01 AI176287, K24 HL166024), the Massachusetts Consortium on Pathogen Readiness, and the Massachusetts General Hospital Department of Medicine.
Keywords: COVID-19; Clinical Research; Infectious disease.
Figures

References
-
- WHO. COVID-19 epidemiological update. https://www.who.int/publications/m/item/covid-19-epidemiological-update-... Updated October 9, 2024.Accessed September 9, 2025.
-
- WHO. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. https://www.who.int/news-room/statements/26-11-2021-classification-of-om... Updtaed November 26, 2021. Accessed September 9, 2025.
-
- CDC. New SARS-CoV-2 variant of concern identified: Omicron (B.1.1.529) variant. https://stacks.cdc.gov/view/cdc/112179 Updated December 1, 2021. Accessed September 9, 2025.
-
- CDC. Preventing Spread of Respiratory Viruses When You’re Sick. https://www.cdc.gov/respiratory-viruses/prevention/precautions-when-sick... Updated August 18, 2025. Accessed September 9, 2025.
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

