Streptococcus pyogenes EVs induce the alternative inflammasome via caspase-4/-5 in human monocytes
- PMID: 40925957
- PMCID: PMC12508482
- DOI: 10.1038/s44319-025-00558-7
Streptococcus pyogenes EVs induce the alternative inflammasome via caspase-4/-5 in human monocytes
Abstract
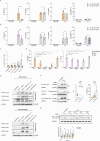
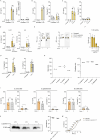
The sensing of Gram-negative Extracellular Vesicles (EVs) by the innate immune system has been extensively studied in the past decade. In contrast, recognition of Gram-positive EVs by innate immune cells remains poorly understood. Comparative genome-wide transcriptional analysis in human monocytes uncovered that S. pyogenes EVs induce proinflammatory signatures that are markedly distinct from those of their parental cells. Among the 209 genes exclusively upregulated by EVs, caspase-5 prompted us to study inflammasome signaling pathways in depth. We show that lipoteichoic acid (LTA), a structural component of Gram-positive bacterial membranes present on EVs from S. pyogenes and other Gram-positive species, is sensed by TLR2 which triggers the alternative inflammasome composed of NLRP3 and the inflammatory caspases-4/-5 to mount an IL-1β response without inducing cell death. For S. pyogenes, we identify TLR8 as a sensor to mediate caspase-4/-5-dependent IL-1β secretion. Notably, inflammasome activation by intact bacteria is independent of the global virulence regulator CovS in monocytes. Overall, our study highlights a new role for TLR2 and caspase-4/-5 in the recognition of Gram-positive EVs in human monocytes.
Keywords: Streptococcus pyogenes; Caspases; Extracellular Vesicles; Inflammasome; Monocytes.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures









References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

