Genetic variance in the murine defensin locus modulates glucose homeostasis
- PMID: 40926119
- PMCID: PMC12528722
- DOI: 10.1038/s44318-025-00555-5
Genetic variance in the murine defensin locus modulates glucose homeostasis
Abstract
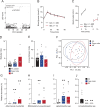
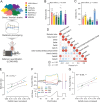
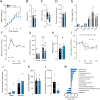
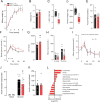
Insulin resistance is a heritable risk factor for many chronic diseases; however, the genetic drivers remain elusive. In seeking these, we performed genetic mapping of insulin sensitivity in 670 chow-fed Diversity Outbred in Australia (DOz) mice and identified a genome-wide significant locus (QTL) on chromosome 8 encompassing 17 defensin genes. By taking a systems genetics approach, we identified alpha-defensin 26 (Defa26) as the causal gene in this region. To validate these findings, we synthesized Defa26 and performed diet supplementation experiments in two mouse strains with distinct endogenous Defa26 expression levels. In the strain with relatively lower endogenous expression (C57BL/6J) supplementation improved insulin sensitivity and reduced gut permeability, while in the strain with higher endogenous expression (A/J) it caused hypoinsulinemia, glucose intolerance and muscle wasting. Based on gut microbiome and plasma bile acid profiling this appeared to be the result of disrupted microbial bile acid metabolism. These data illustrate the danger of single strain over-reliance and provide the first evidence of a link between host-genetics and insulin sensitivity which is mediated by the microbiome.
Keywords: Bile Acids; Defensin; Diversity Outbred; Insulin Sensitivity; Microbiome.
© 2025. Crown.
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

