Whole genome sequence analysis of low-density lipoprotein cholesterol across 246 K individuals
- PMID: 40926209
- PMCID: PMC12418676
- DOI: 10.1186/s13059-025-03698-0
Whole genome sequence analysis of low-density lipoprotein cholesterol across 246 K individuals
Abstract
Background: Rare genetic variation provided by whole genome sequence datasets has been relatively less explored for its contributions to human traits. Meta-analysis of sequencing data offers advantages by integrating larger sample sizes from diverse cohorts, thereby increasing the likelihood of discovering novel insights into complex traits. Furthermore, emerging methods in genome-wide rare variant association testing further improve power and interpretability.
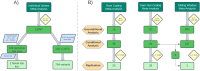
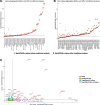
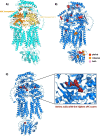
Results: Here, we conduct the largest meta-analysis of whole genome sequencing for low-density lipoprotein cholesterol (LDL-C), a therapeutic target for coronary artery disease, analyzing data from 246 K participants and integrating 1.23B variants from the UK Biobank and the Trans-Omics for Precision Medicine (TOPMed) program. We identify numerous rare coding and non-coding gene associations related to LDL-C, with replication across 86 K participants in All of Us. Our findings are based on single-variant analyses, rare coding and non-coding variant aggregation tests, and sliding window approaches. Through this comprehensive analysis, we identify 704 novel single-variant associations, 25 novel rare coding variant aggregates, 28 novel rare non-coding variant aggregates, and one novel sliding window aggregate.
Conclusions: This study provides a meta-analysis framework for large-scale whole genome sequence association analyses from diverse population groups, yielding novel rare non-coding variant associations.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: For the TOPMed cohort, study participants provided consent per each study’s Institutional Review Board (IRB)-approved protocol. The TOPMed data analysis is associated with paper proposal ID 15536. For UKB participants, written informed consent was given per the UKB primary protocol. UK Biobank data analysis was facilitated through UKB application 7089. For the AOU cohort, written informed consent was provided in accordance with the primary Institutional Review Board for AOU. AOU data analysis was facilitated through the AOU Researcher Workbench. Secondary use of UK Biobank, TOPMed, and AOU data was approved by the Massachusetts Hospital Institutional Review Board. Consent for publication: All participants provided informed consent for publication. Competing interests: P.N. reports research grants from Allelica, Amgen, Apple, Boston Scientific, Cleerly, Genentech / Roche, Ionis, Novartis, and Silence Therapeutics, personal fees from AIRNA, Allelica, Apple, AstraZeneca, Bain Capital, Blackstone Life Sciences, Bristol Myers Squibb, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co, Esperion Therapeutics, Foresite Capital, Foresite Labs, Genentech / Roche, GV, HeartFlow, Magnet Biomedicine, Merck, Novartis, Novo Nordisk, TenSixteen Bio, and Tourmaline Bio, equity in Bolt, Candela, Mercury, MyOme, Parameter Health, Preciseli, and TenSixteen Bio, royalties from Recora for intensive cardiac rehabilitation, and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. The remaining authors declare no competing interests.
Figures
References
-
- Tsao CW, Aday AW, Almarzooq ZI, Alonso A, Beaton AZ, Bittencourt MS, et al. Heart disease and stroke statistics—2022 update: A report from the American Heart Association. Circulation 2022 Feb 22 [cited 2024 Mar 13];145(8). Available from: https://pubmed.ncbi.nlm.nih.gov/35078371/. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

