Dysregulated RNA splicing impairs regeneration in alcohol-associated liver disease
- PMID: 40931030
- PMCID: PMC12423296
- DOI: 10.1038/s41467-025-63251-2
Dysregulated RNA splicing impairs regeneration in alcohol-associated liver disease
Abstract
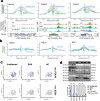
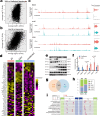
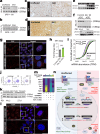
Individuals with progressive liver failure risk dying without liver transplantation. However, our understanding of why regenerative responses are disrupted in failing livers is limited. Here, we perform multiomic profiling of healthy and diseased human livers using bulk and single-nucleus RNA- and ATAC-seq. We report that in alcohol-associated liver disease, alterations in the hepatic immune milieu prevent hepatocytes from transitioning to proliferative progenitors. We also find differences in RNA binding protein expression, particularly of the ESRP, PTBP, and SR families, leading to misregulation of developmentally controlled RNA splicing. Our data pinpoint ESRP2 as a disease-sensitive splicing factor and support a causal role for its deficiency in the pathogenesis of severe alcoholic hepatitis. Notably, splicing defects in Tcf4 and Slk, two ESPR2 targets, alter their nuclear localization and activities, disrupting WNT and Hippo signaling pathways that are critical for normal liver regeneration. We further demonstrate that changes in stromal cell populations enrich failing livers with TGF-β, which suppresses the ESRP2-driven epithelial splicing program and replaces functional parenchyma with quasi-progenitor-like cells lacking liver-specific functions. Taken together, these findings indicate that misspliced RNAs are effective biomarkers for alcohol-associated liver disease, and targeting them could improve recovery in affected individuals.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Update of
-
Dysregulated RNA splicing induces regeneration failure in alcohol-associated liver disease.bioRxiv [Preprint]. 2024 Nov 30:2024.11.29.626099. doi: 10.1101/2024.11.29.626099. bioRxiv. 2024. Update in: Nat Commun. 2025 Sep 10;16(1):8049. doi: 10.1038/s41467-025-63251-2. PMID: 39651310 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01HL126845/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R21 HD104039/HD/NICHD NIH HHS/United States
- R21HD104039/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- R01 DK077794/DK/NIDDK NIH HHS/United States
- R01 HL126845/HL/NHLBI NIH HHS/United States
- MDA1072487/Muscular Dystrophy Association (Muscular Dystrophy Association Inc.)
- P30 AI117943/AI/NIAID NIH HHS/United States
- R01 AA010154/AA/NIAAA NIH HHS/United States
- R01AA010154/U.S. Department of Health & Human Services | NIH | National Institute on Alcohol Abuse and Alcoholism (NIAAA)
- R24 AA025017/AA/NIAAA NIH HHS/United States
- T32 EB019944/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

