PATJ deficiency leads to cystic kidney disease and related ciliopathies
- PMID: 40931526
- PMCID: PMC12512994
- DOI: 10.1016/j.xhgg.2025.100514
PATJ deficiency leads to cystic kidney disease and related ciliopathies
Abstract
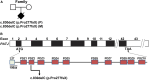
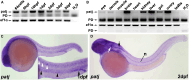
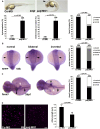
Cystic kidney disease and related ciliopathies are caused by pathogenic variants in genes that commonly result in ciliary dysfunction. For a substantial number of individuals affected by those cilia-related diseases, the causative gene remains unknown. Using massively parallel sequencing, we here identified a pathogenic bi-allelic variant in the gene encoding PALS1-associated tight junction protein ([PATJ] also known as inactivation-no-afterpotential D-like, INADL) in an individual with ciliopathy. The affected fetus carried the homozygous truncating PATJ nonsense variant c.830delC (p.Pro277fsX), and presented with a syndromic phenotype mainly characterized by polycystic kidney disease and hydrocephalus. Using zebrafish (Danio rerio) as a vertebrate in vivo model organism, we could validate our patient findings and demonstrated a ciliopathy phenotype. In addition, we were able to address a hitherto not described role of Patj for cilia formation and function. Taken together, with the Crumbs cell polarity complex member PATJ, we add a new member to the large family of ciliopathy-related human disease proteins that is different from the classical ciliopathy protein classes, and may offer new perspectives for drug development.
Keywords: MPDZ; MUPP1; Reissner fiber; cilia; ciliopathy; hydrocephalus; massively parallel sequencing; morpholino; polycystic kidney disease; zebrafish.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
LinkOut - more resources
Full Text Sources

