Banana breeding by genome design
- PMID: 40932063
- PMCID: PMC12590350
- DOI: 10.1111/jipb.70025
Banana breeding by genome design
Abstract
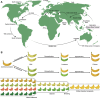
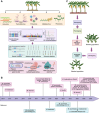
Bananas and plantains of the genus Musa constitute the most vital fruits and staple foods. Cultivated bananas may have originated from intraspecific and interspecific hybridizations of four wild species, namely Musa acuminata (A), M. balbisiana (B), M. schizocarpa (S), and the Australimusa species (T). Here, we appraise the advances made in banana genomics, genetics, and breeding over the past few decades. The sequencing of Musa genomes has been a major breakthrough in banana research programs, presenting unprecedented possibilities for gaining deeper insights into the evolution, domestication, breeding, and genetics of indispensable agronomic traits of bananas. Also, we delve into how these genetic facets, coupled with innovative genomic-assisted tools, including genomic selection and gene editing, propel advancements in banana breeding endeavors. Ultimately, we propose the forthcoming prospects within the domain of banana genetics and breeding.
Keywords: Musa; bananas; domestication; genetics; genomic breeding; genomics.
© 2025 The Author(s). Journal of Integrative Plant Biology published by John Wiley & Sons Australia, Ltd on behalf of Institute of Botany, Chinese Academy of Sciences.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



References
-
- Álvarez‐López, D. , Herrera‐Valencia, V.A. , Góngora‐Castillo, E. , García‐Laynes, S. , Puch‐Hau, C. , López‐Ochoa, L.A. , Lizama‐Uc, G. , and Peraza‐Echeverria, S. (2022). Genome‐wide analysis of the LRR‐RLP gene family in a wild banana (Musa acuminata ssp. malaccensis) uncovers multiple Fusarium wilt resistance gene candidates. Genes 13: 638. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- CARS-31/the China Agriculture Research System
- CATASCXTD202309/Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences
- U22A20487/National Natural Science Foundation of China
- NKLTCB202306/the project of the National Key Laboratory for Tropical Crop Breeding
- the Science Fund Program for Distinguished Young Scholars of the National Natural Science Foundation of China (Overseas) to Yongfeng Zhou
LinkOut - more resources
Full Text Sources
Miscellaneous

