Deciphering differences in DNA methylation and transcriptome profiles of oocytes from pigs with high and low developmental competence
- PMID: 40933923
- PMCID: PMC12418950
- DOI: 10.1093/eep/dvaf018
Deciphering differences in DNA methylation and transcriptome profiles of oocytes from pigs with high and low developmental competence
Abstract
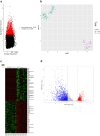
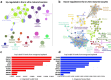
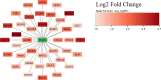
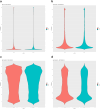
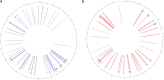
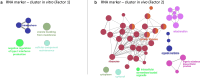
In vitro maturation (IVM) is a critical step in animal in vitro embryo production, yet oocytes matured in vitro often exhibit lower developmental competence than their in vivo counterparts. However, the molecular mechanisms behind this observation remain unclear. This study investigated the gene expression and DNA methylation profiles in porcine oocytes with different developmental competencies. To study these differences, we used as a model oocytes from prepubertal gilts (IVM) and sows (in vivo matured) and assessed their developmental competence up to the blastocyst stage. We also examined their gene expression and DNA methylation profiles at single-cell resolution using RNA sequencing and bisulfite sequencing. Oocytes were obtained by aspiration of either ovarian follicles between 3 and 6 mm diameter, and the subsequent IVM, or ovarian follicles from 8 to 10 mm diameter, with no need for maturation (in vivo matured oocytes). Cleavage rates (58.2 ± 3.0 and 45.7 ± 4.4) and blastocyst rates (31.4 ± 3.7 and 47.5 ± 6.6) for IVM and in vivo groups differed significantly. Using the in vivo group as a reference, IVM oocytes had 1297 downregulated and 476 upregulated differentially expressed genes (DEGs), with upregulated DEGs associated with organelle organization and cell cycle processes, and downregulated genes involved in protein synthesis and metabolomic processes. While global DNA methylation levels were similar between groups, a few differentially methylated regions were found in CpG islands, promoters, and coding regions. Our integrative analysis identified key methylated regions and genes that distinguish each group, suggesting that both donor age and maturation conditions significantly influence gene expression regulation in oocytes with different developmental competencies.
Keywords: cellular division; epigenomic; multi-omic analysis; oocyte maturation; porcine.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures

References
-
- Thomson KJ. World agriculture: towards 2015/2030: an FAO perspective. Land Use Policy. 2003;20:375.
-
- Tilman D, Cassman KG, Matson PA, et al. Agricultural sustainability and intensive production practices. Nature. 2002;418:671–77. - PubMed
-
- Mattioli M, Bacci ML, Galeati G, et al. Developmental competence of pig oocytes matured and fertilized in vitro. Theriogenology. 1989;31:1201–1207. - PubMed
-
- Chen PR, Uh K, Redel BK, et al. Production of pigs from porcine embryos generated in vitro. Front Anim Sci. 2022;3:1–13.
-
- Grupen CG. The evolution of porcine embryo in vitro production. Theriogenology. 2014;81:24–37. - PubMed
LinkOut - more resources
Full Text Sources

