Cell Type-Specific Contributions of UBE3A to Angelman Syndrome Behavioral Phenotypes
- PMID: 40935670
- PMCID: PMC12479163
- DOI: 10.1523/ENEURO.0453-24.2025
Cell Type-Specific Contributions of UBE3A to Angelman Syndrome Behavioral Phenotypes
Abstract
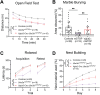
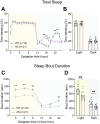
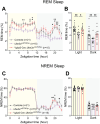
Angelman syndrome (AS) is a rare neurodevelopmental disorder caused by loss of expression of the maternal UBE3A allele and is characterized by a constellation of impactful neurologic symptoms. While previous work has uncovered outsized contributions of GABAergic neuron-selective Ube3a deletion to seizure susceptibility and electroencephalography (EEG) phenotypes in a mouse model of AS, the neuronal populations governing a broader range of behaviors have not been studied. Here, we used male and female mice to test the consequences of Ube3a deletion from GABAergic or glutamatergic neurons across a well-characterized battery of AS-relevant behaviors. Surprisingly, we observed deficits in numerous motor and innate behaviors in mice with glutamatergic Ube3a deletion and relatively few consequences of GABAergic Ube3a deletion. Furthermore, genetic Ube3a reinstatement in glutamatergic neurons rescued multiple motor and innate behaviors. When tested for sleep-wake behaviors, the selective loss of Ube3a from glutamatergic neurons disrupted sleep similarly to that of AS model mice (Ube3am-/p+ ), and glutamatergic Ube3a reinstatement overcame the lack of active cycle "siesta" and decreased REM phenotypes observed in AS model mice. Altogether, this work demonstrates a major role of glutamatergic neuron UBE3A loss in mediating multiple AS behavioral features, suggesting a divergence from the circuitry underlying enhanced seizure susceptibility. Our findings imply that neuronal cell type-agnostic UBE3A reinstatement is likely required for successful AS genetic therapies-with reinstatement of UBE3A in GABAergic neurons necessary for overcoming epileptic and EEG phenotypes, and reinstatement in glutamatergic neurons necessary for overcoming most other behavioral phenotypes.
Keywords: Angelman; behavior; mouse; neurodevelopment; sleep; ube3a.
Copyright © 2025 Ringelberg et al.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
