CDK4/6 Inhibitor Priming Enhances PD-1 Blockade via Sellhi Neutrophil-Induced Stat5a+ Progenitor Exhausted CD8+ T Cell
- PMID: 40936164
- PMCID: PMC12667468
- DOI: 10.1002/advs.202510501
CDK4/6 Inhibitor Priming Enhances PD-1 Blockade via Sellhi Neutrophil-Induced Stat5a+ Progenitor Exhausted CD8+ T Cell
Abstract
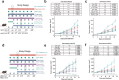
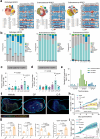
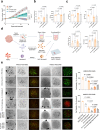
Cell cycle pathway, especially via cyclin D1-CDK4/6 signaling, is enriched in immunotherapy-resistant and immune-excluded tumors. CDK4/6 inhibitor (CDK4/6i) induces antitumor immune phenotypes by targeting both tumor and immune cells, enhancing immune checkpoint blockade (ICB), but optimal combination modalities and the corresponding cellular mechanisms remain unclear. Here, it is shown that activation of tumor cell-intrinsic cyclin D1-CDK4/6 signaling is associated with low tumor-infiltrating lymphocyte populations and immunotherapy resistance in head and neck squamous cell carcinoma (HNSCC). Comparison of sequential versus combinatorial regimens in subcutaneous or orthotopic HNSCC mice revealed that CDK4/6i priming before anti-PD-1 enhances response durability by promoting CD8+ and CD4+ T cell infiltration and decreasing overall neutrophil abundance. Mechanistically, IL15-secreted Sell(hi) neutrophils induced Stat5a+ progenitor exhausted CD8+ T cells contributed to the antitumor effect of CDK4/6i priming modalities. Together with corroborating evidence from a clinically relevant patient-derived-organoid-TIL (PDO-TIL) co-culture model, these findings support further clinical testing of brief CDK4/6i dosing before anti-PD-1 to improve ICB efficacy.
Keywords: CD8+Tpex; CDK4/6i priming; HNSCC; Sell(hi) neutrophils; Stat5a.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- a) Burtness B., Harrington K. J., Greil R., Soulières D., Tahara M., de Castro G. Jr., Psyrri A., Basté N., Neupane P., Bratland Å., Fuereder T., Hughes B. G. M., Mesía R., Ngamphaiboon N., Rordorf T., Wan Ishak W. Z., Hong R. L., González Mendoza R., Roy A., Zhang Y., Gumuscu B., Cheng J. D., Jin F., Rischin D., Lancet 2019, 394, 1915;
- b) Jin Z., Zhang B., Zhang L., Huang W., Mo X., Chen Q., Wang F., Chen Z., Li M., Zhang S., Ther. Adv. Med. Oncol. 2020, 12, 1758835920983717. - PMC - PubMed
-
- a) Spranger S., Bao R., Gajewski T. F., Nature 2015, 523, 231; - PubMed
- b) Peng W., Chen J. Q., Liu C., Malu S., Creasy C., Tetzlaff M. T., Xu C., McKenzie J. A., Zhang C., Liang X., Williams L. J., Deng W., Chen G., Mbofung R., Lazar A. J., Torres‐Cabala C. A., Cooper Z. A., Chen P. L., Tieu T. N., Spranger S., Yu X., Bernatchez C., Forget M. A., Haymaker C., Amaria R., McQuade J. L., Glitza I. C., Cascone T., Li H. S., Kwong L. N., et al., Cancer Discov. 2016, 6, 202; - PMC - PubMed
- c) Hamarsheh S., Groß O., Brummer T., Zeiser R., Nat. Commun. 2020, 11, 5439. - PMC - PubMed
-
- Jerby‐Arnon L., Shah P., Cuoco M. S., Rodman C., Su M. J., Melms J. C., Leeson R., Kanodia A., Mei S., Lin J. R., Wang S., Rabasha B., Liu D., Zhang G., Margolais C., Ashenberg O., Ott P. A., Buchbinder E. I., Haq R., Hodi F. S., Boland G. M., Sullivan R. J., Frederick D. T., Miao B., Moll T., Flaherty K. T., Herlyn M., Jenkins R. W., Thummalapalli R., Kowalczyk M. S., et al., Cell 2018, 175, 984. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
