Matrix directs trophoblast differentiation in a bioprinted organoid model of early placental development
- PMID: 40940337
- PMCID: PMC12432263
- DOI: 10.1038/s41467-025-62996-0
Matrix directs trophoblast differentiation in a bioprinted organoid model of early placental development
Abstract
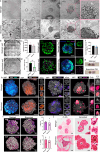
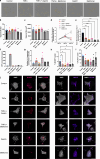
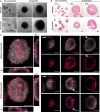
Trophoblast organoids can provide crucial insights into mechanisms of placentation, however their potential is limited by highly variable extracellular matrices unable to reflect in vivo tissues. Here, we present a bioprinted placental organoid model, generated using the first trimester trophoblast cell line, ACH-3P, and a synthetic polyethylene glycol (PEG) matrix. Bioprinted or Matrigel-embedded organoids differentiate spontaneously from cytotrophoblasts into two major subtypes: extravillous trophoblasts (EVTs) and syncytiotrophoblasts (STBs). Bioprinted organoids are driven towards EVT differentiation and show close similarity with early human placenta or primary trophoblast organoids. Inflammation inhibits proliferation and STBs within bioprinted organoids, which aspirin or metformin (0.5 mM) cannot rescue. We reverse the inside-out architecture of ACH-3P organoids by suspension culture with STBs forming on the outer layer of organoids, reflecting placental tissue. Our bioprinted methodology is applicable to trophoblast stem cells. We present a high-throughput, automated, and tuneable trophoblast organoid model that reproducibly mimics the placental microenvironment in health and disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.V. was an employee of Inventia Life Science at the time of the study. The remaining authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

