Genetic subtyping of obesity reveals biological insights into the uncoupling of adiposity from its cardiometabolic comorbidities
- PMID: 40940440
- PMCID: PMC12618246
- DOI: 10.1038/s41591-025-03931-0
Genetic subtyping of obesity reveals biological insights into the uncoupling of adiposity from its cardiometabolic comorbidities
Abstract
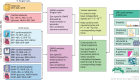
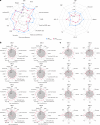
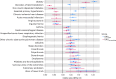
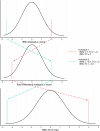
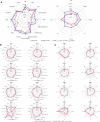
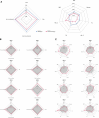
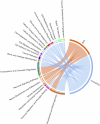
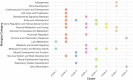
Obesity is a heterogeneous condition not adequately captured by a single adiposity trait. We conducted a multi-trait genome-wide association analysis using individual-level data from 452,768 UK Biobank participants to study obesity in relation to cardiometabolic health. We defined continuous 'uncoupling phenotypes', ranging from high adiposity with healthy cardiometabolic profiles to low adiposity with unhealthy ones. We identified 266 variants across 205 genomic loci where adiposity-increasing alleles were simultaneously associated with lower cardiometabolic risk. A genetic risk score (GRSuncoupling) aggregating these variants was associated with a lower risk of cardiometabolic disorders, including dyslipidemia and ischemic heart disease, despite higher obesity risk; unlike an adiposity score based on body fat percentage-associated variants (GRSBFP). The 266 variants formed eight genetic subtypes of obesity, each with distinct risk profiles and pathway signatures. Proteomic analyses revealed signatures separating adiposity- and health-driven effects. Our findings reveal new mechanisms that uncouple obesity from cardiometabolic comorbidities and lay a foundation for genetically informed subtyping of obesity to support precision medicine.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.C.H. has received honoraria for expert roles from Novo Nordisk and Rhythm Pharmaceuticals and provides training and treatment of obesity. All the other authors declare no competing interests.
Figures





Update of
-
Genetic subtyping of obesity reveals biological insights into the uncoupling of adiposity from its cardiometabolic comorbidities.medRxiv [Preprint]. 2025 Feb 28:2025.02.25.25322830. doi: 10.1101/2025.02.25.25322830. medRxiv. 2025. Update in: Nat Med. 2025 Nov;31(11):3801-3812. doi: 10.1038/s41591-025-03931-0. PMID: 40061343 Free PMC article. Updated. Preprint.
References
-
- Mokdad, A. H. et al. Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA289, 76–79 (2003). - PubMed
-
- Alberti, K. G. et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation120, 1640–1645 (2009). - PubMed
-
- Hubert, H. B., Feinleib, M., McNamara, P. M. & Castelli, W. P. Obesity as an independent risk factor for cardiovascular disease: a 26-year follow-up of participants in the Framingham Heart Study. Circulation67, 968–977 (1983). - PubMed
-
- Must, A. et al. The disease burden associated with overweight and obesity. JAMA282, 1523–1529 (1999). - PubMed
MeSH terms
Grants and funding
- U01 HG007417/HG/NHGRI NIH HHS/United States
- R01 HL158884/HL/NHLBI NIH HHS/United States
- 75N92022D00002/HL/NHLBI NIH HHS/United States
- R01 HL163262/HL/NHLBI NIH HHS/United States
- NNF22SA0079901/Novo Nordisk Fonden (Novo Nordisk Foundation)
- R01 DK107786/DK/NIDDK NIH HHS/United States
- R01 DK075787/DK/NIDDK NIH HHS/United States
- S10 OD026880/OD/NIH HHS/United States
- R01 DK123019/DK/NIDDK NIH HHS/United States
- R01 HL151152/HL/NHLBI NIH HHS/United States
- 75N92022D00003/HL/NHLBI NIH HHS/United States
- R01DK110113/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- UM1 DK126185/DK/NIDDK NIH HHS/United States
- R01 DK122503/DK/NIDDK NIH HHS/United States
- R01 HL142302/HL/NHLBI NIH HHS/United States
- R01 DK110113/DK/NIDDK NIH HHS/United States
- 75N92022D00004/HL/NHLBI NIH HHS/United States
- R01 DK102173/DK/NIDDK NIH HHS/United States
- U01 HG011723/HG/NHGRI NIH HHS/United States
- R01 HL156991/HL/NHLBI NIH HHS/United States
- R01 DK139598/DK/NIDDK NIH HHS/United States
- UL1 TR004419/TR/NCATS NIH HHS/United States
- S10 OD030463/OD/NIH HHS/United States
- 75N92022D00005/HL/NHLBI NIH HHS/United States
- R35 HL155466/HL/NHLBI NIH HHS/United States
- R01 HG010297/HG/NHGRI NIH HHS/United States
- 75N92022D00001/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

