CDK1-mediated phosphorylation of LDHA fuels mitosis through LDHB-dependent lactate oxidation
- PMID: 40940446
- PMCID: PMC12550033
- DOI: 10.1038/s44319-025-00573-8
CDK1-mediated phosphorylation of LDHA fuels mitosis through LDHB-dependent lactate oxidation
Abstract
While cancer cells overexpress lactate dehydrogenase A (LDHA) to support glycolytic flux and lactate production, the role of LDHB-which preferentially catalyzes lactate oxidation-remains unclear. Here, we demonstrate that LDHB, but not LDHA, is essential for mitotic progression in cancers. During mitosis, CDK1 phosphorylates LDHA at threonine 18, reducing its incorporation into the lactate dehydrogenase (LDH) tetramer. This results in LDHB-enriched tetramers that shift catalytic activity toward lactate oxidation, converting lactate and NAD⁺ into pyruvate and NADH. The generated NADH fuels oxidative phosphorylation and ATP production, thereby sustaining mitosis. Notably, LDHA-T18 phosphorylation occurs exclusively in tumor tissues. Our findings reveal a tumor-specific mechanism in which CDK1 reprograms LDH isoenzyme composition to direct lactate toward NADH production, ensuring energy homeostasis during mitosis. This underscores the therapeutic necessity of targeting both LDHA and LDHB in cancer.
Keywords: ATP; Lactate; Lactate Dehydrogenase; Mitosis; NADH.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures


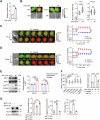

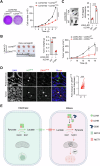


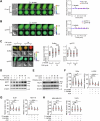
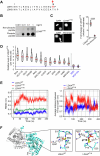

References
MeSH terms
Substances
Grants and funding
- 31970670/MOST | National Natural Science Foundation of China (NSFC)
- 32170736/MOST | National Natural Science Foundation of China (NSFC)
- 92357301/MOST | National Natural Science Foundation of China (NSFC)
- XDB0940101/The strategic priority Research Program of the Chinese academy of sciences
- YD9100002028/the Fundamental Research Funds for the Central Universities
LinkOut - more resources
Full Text Sources
Miscellaneous

