Computational design of sequence-specific DNA-binding proteins
- PMID: 40940539
- PMCID: PMC12618268
- DOI: 10.1038/s41594-025-01669-4
Computational design of sequence-specific DNA-binding proteins
Abstract
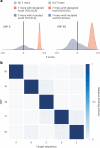
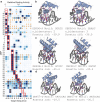
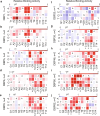
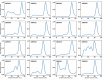
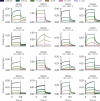
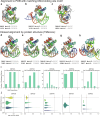
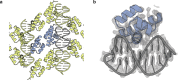
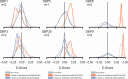
Sequence-specific DNA-binding proteins (DBPs) have critical roles in biology and biotechnology and there has been considerable interest in the engineering of DBPs with new or altered specificities for genome editing and other applications. While there has been some success in reprogramming naturally occurring DBPs using selection methods, the computational design of new DBPs that recognize arbitrary target sites remains an outstanding challenge. We describe a computational method for the design of small DBPs that recognize short specific target sequences through interactions with bases in the major groove and use this method to generate binders for five distinct DNA targets with mid-nanomolar to high-nanomolar affinities. The individual binding modules have specificity closely matching the computational models at as many as six base-pair positions and higher-order specificity can be achieved by rigidly positioning the binders along the DNA double helix using RFdiffusion. The crystal structure of a designed DBP-target site complex is in close agreement with the design model and the designed DBPs function in both Escherichia coli and mammalian cells to repress and activate transcription of neighboring genes. Our method provides a route to small and, hence, readily deliverable sequence-specific DBPs for gene regulation and editing.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: C.G., R.P., R.M., C.N., F.D., D.C., B.C., H.H., G-R.L. and D.B. are coinventors on a provisional patent application that incorporates discoveries described in this paper. The other authors declare no competing interests.
Figures







Update of
-
Computational design of sequence-specific DNA-binding proteins.bioRxiv [Preprint]. 2023 Sep 21:2023.09.20.558720. doi: 10.1101/2023.09.20.558720. bioRxiv. 2023. Update in: Nat Struct Mol Biol. 2025 Nov;32(11):2252-2261. doi: 10.1038/s41594-025-01669-4. PMID: 37790440 Free PMC article. Updated. Preprint.
References
-
- Wilken, M. S. et al. Quantitative dialing of gene expression via precision targeting of KRAB repressor. Preprint at bioRxiv10.1101/2020.02.19.956730 (2020).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

