An Update on Single-Cell RNA Sequencing in Illuminating Disease Mechanisms of Cutaneous T-Cell Lymphoma
- PMID: 40941016
- PMCID: PMC12427944
- DOI: 10.3390/cancers17172921
An Update on Single-Cell RNA Sequencing in Illuminating Disease Mechanisms of Cutaneous T-Cell Lymphoma
Abstract
Cutaneous T-cell Lymphomas (CTCLs) are a heterogeneous group of non-Hodgkin lymphomas that currently have an incompletely understood pathophysiology and several challenges in both diagnosis and management. Single-cell RNA sequencing (scRNA-seq) is a powerful tool that enables the analysis of gene expression at the individual-cell level, revealing cellular heterogeneity and a complex tumor microenvironment. As single-cell RNA sequencing has become increasingly utilized, we aimed to provide an update on recent notable applications of single-cell RNA sequencing in CTCL and their findings. The included studies highlight the intricate network of interactions in the tumor microenvironment that contributes to tumorigenesis. While CTCL is notoriously heterogeneous, our results identify key markers that prove promising for diagnosis, prognostication, and therapeutic targets.
Keywords: CTCL; Sézary syndrome; biomarkers; cutaneous T-cell lymphoma; genomic; mycosis fungoides; scRNA-seq; single-cell RNA sequencing; transcriptomic; tumor microenvironment.
Conflict of interest statement
LJG has served as an investigator for and/or received research support from Helsinn Group, J&J, Mallinckrodt, Kyowa Kirin, Soligenix, Innate, Merck, BMS, and Stratpharma; on the speakers’ bureau for Helsinn Group and J&J; and on the scientific advisory board for Helsinn Group, J&J, Mallinckrodt, Sanofi, Regeneron, and Kyowa Kirin. The other authors have no conflicts of interest to declare.
Figures


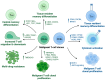


References
-
- Campo E., Jaffe E.S., Cook J.R., Quintanilla-Martinez L., Swerdlow S.H., Anderson K.C., Brousset P., Cerroni L., de Leval L., Dirnhofer S., et al. The International Consensus Classification of Mature Lymphoid Neoplasms: A report from the Clinical Advisory Committee. Blood. 2022;140:1229–1253. doi: 10.1182/blood.2022015851. - DOI - PMC - PubMed
-
- Alaggio R., Amador C., Anagnostopoulos I., Attygalle A.D., Araujo I.B.O., Berti E., Bhagat G., Borges A.M., Boyer D., Calaminici M., et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia. 2022;36:1720–1748. doi: 10.1038/s41375-022-01620-2. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

