Integrated Multi-Omics Analysis Reveals the Role of Resveratrol in Regulating the Intestinal Function of Megalobrama amblycephala via m6A Methylation
- PMID: 40943507
- PMCID: PMC12429812
- DOI: 10.3390/ijms26178587
Integrated Multi-Omics Analysis Reveals the Role of Resveratrol in Regulating the Intestinal Function of Megalobrama amblycephala via m6A Methylation
Abstract
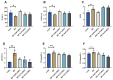
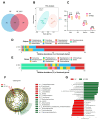
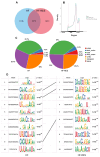
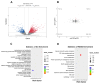
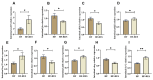
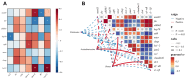
Resveratrol (RES), a natural polyphenol with lipid metabolism-regulating properties, also demonstrates remarkable efficacy in strengthening intestinal barrier integrity. In order to elucidate the mechanism by which RES ameliorates intestinal damage and lipid metabolism disturbances in Megalobrama amblycephala under a high-fat (HF) diet, a conventional diet (CON), an HF diet (HF), or an HF diet supplemented with 0.6, 3, or 6 g/kg RES (HF + 0.06%, 0.3%, or 0.6% RES) was fed to fish. After 8 weeks, RES supplementation in the HF diet significantly improved the growth performance and alleviated hepatic lipid deposition. Microbiota profiling revealed RES improved intestinal barrier function by reducing α-diversity, Actinobacteria and Bosea abundances, and enriching Firmicutes abundance. RES also maintained the integrity of the intestinal physical barrier and inhibited the inflammatory response. MeRIP-seq analysis indicated that RES modulated intestinal mRNA m6A methylation by upregulating methyltransferase-like 3 (mettl3) and downregulating fat mass and obesity-associated gene (fto) and Alk B homolog 5 (alkbh5). Combined RNA-seq and MeRIP-seq data revealed that RES alleviated endoplasmic reticulum stress (ERS) by upregulating the m6A methylation and gene level of heat shock protein 70 (hsp70). Correlation analyses identified significant associations between intestinal microbiota composition and ERS, tight junction, and inflammation. In summary, RES ameliorates lipid dysregulation via a synergistic mechanism involving intestinal microbiota, m6A modification, ERS, barrier function, and inflammatory response.
Keywords: endoplasmic reticulum stress; high lipid metabolism; intestinal barrier function; m6A methylation; resveratrol.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Fu S., Xie X., Zhang W., Cao Z. The nuntrition of Silurus meridionalis: III. protein-sparing effect og dietary lipid. Acta Hydrobiol. Sin. 2001;25:70–75. doi: 10.3724/issn1000-3207-2001-1-70-3. - DOI
-
- Lu K., Xu W., Li J., Li X., Huang G., Liu W. Alterations of liver histology and blood biochemistry in blunt snout bream Megalobrama amblycephala fed high-fat diets. Fish. Sci. 2013;79:661–671. doi: 10.1007/s12562-013-0635-4. - DOI
-
- Wei M., Song L., Yuan X., Li H., Ji H., Sun J. Dietary supplementation with a PPARγ agonist promotes adipocyte hyperplasia and improves high-fat diet tolerance and utilization in grass carp (Ctenopharyngodon idellus) Aquaculture. 2023;578:740081. doi: 10.1016/j.aquaculture.2023.740081. - DOI
-
- Wei M., Yuan X., Song L., Li H., Ji H., Sun J. Comparative proteomic analysis of pathological characterization of adipose tissue remodeling induced by high-fat diet and high-carbohydrate diet in grass carp (Ctenopharyngodon idellus) Aquaculture. 2024;590:741079. doi: 10.1016/j.aquaculture.2024.741079. - DOI
-
- Yu C., Zhang J., Qin Q., Liu J., Xu J., Xu W. Berberine improved intestinal barrier function by modulating the intestinal microbiota in blunt snout bream (Megalobrama amblycephala) under dietary high-fat and high-carbohydrate stress. Fish Shellfish Immunol. 2020;102:336–349. doi: 10.1016/j.fsi.2020.04.052. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

