Harmonizing genotype array data to understand genetic risk for brain amyloid burden in the AMYPAD PNHS Consortium
- PMID: 40947441
- PMCID: PMC12433760
- DOI: 10.1002/alz.70376
Harmonizing genotype array data to understand genetic risk for brain amyloid burden in the AMYPAD PNHS Consortium
Abstract
Introduction: We sought to harmonize genotype data from the predementia AMYPAD (Amyloid Imaging to Prevent Alzheimer's Disease) Consortium, compute polygenic risk scores (PRS), and determine their association with global amyloid deposition.
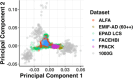
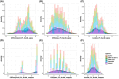
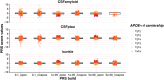
Methods: Genetic data from five AMYPAD parent cohorts were harmonized, and PRS were computed for Alzheimer's disease (AD) susceptibility, cerebrospinal fluid (CSF) amyloid beta (Aβ)42, and CSF phosphorylated tau181. Cross-sectional amyloid (Centiloid [CL]) burden was available for all participants, and regression models determined if PRS were associated with CL burden.
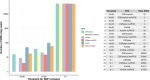
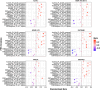
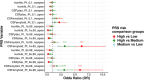
Results: After harmonization, data for 867 participants showed that high CL burden was most strongly predicted by CSF Aβ42 PRS compared to traditional AD susceptibility PRS.
Discussion: This work emphasizes the importance of data harmonization and pooling of cohorts for large-powered studies. Findings suggest a genetic predisposition to amyloid pathology that may predispose individuals early in the AD continuum. This validates the potential use of PRS in clinical (trial) settings as a non-invasive tool to assess AD risk.
Highlights: We developed a robust harmonization pipeline for multi-cohort genotype array data. Cerebrospinal fluid amyloid beta (Aβ)-specific polygenic risk scores (PRS) more strongly predicted global Aβ positron emission tomography burden than other PRS. Results suggest a strong genetic predisposition to early Aβ pathology. This work highlights the need for robust data harmonization and data pooling. This work also validates the potential use of PRS as a non-invasive tool to assess Alzheimer's disease risk.
Keywords: Alzheimer's disease; Amyloid Imaging to Prevent Alzheimer's Disease; amyloid; genotype data harmonization; polygenic risk scores; predementia.
© 2025 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
This communication reflects the views of the authors and neither IMI nor the European Union and EFPIA are liable for any use that may be made of the information contained herein. Research of Alzheimer Center Amsterdam is part of the neurodegeneration research program of Amsterdam Neuroscience. Alzheimer Center Amsterdam is supported by Stichting Alzheimer Nederland and Stichting Steun Alzheimercentrum Amsterdam. EMIF‐Twins‐60+ is supported by the EU/EFPIA Innovative Medicines Initiative Joint Undertaking EMIF grant agreement no. 115372, Alzheimer Nederland and Stichting Dioraphte. P.J.V. is a coinventor on a patent of CSF proteomic subtypes (published under patent no. US2022196683A1, owner VUmc Foundation). C.R. is the founder, CEO, and majority shareholder of Scottish Brain Sciences; has received compensation for study‐related activities from AC Immune SA (paid to institution); has received consulting fees from Biogen, Eisai, MSD, Actinogen, Roche, Eli Lilly, and Novo Nordisk; has received payment or honoraria from Roche, Eisai, and Eli Lilly; and participates on a data safety monitoring board for Novo Nordisk. M.B. is an employee of the Ace Alzheimer Center and an advisory board member for Grifols, Roche, Eli Lilly, Araclon Biotech, Merck, Zambon, Biogen, Novo Nordisk, Bioiberica, Eisai, Servier, and Schwabe Pharma. J.D.G. has received research support from GE HealthCare, Roche Diagnostics, Hoffmann La Roche, and Life‐MI; has participated in symposia sponsored by Biogen, Philips Nederlands, Life‐MI, and Esteve; acted as a consultant for Roche Diagnostics; and served on the molecular neuroimaging advisory board of Prothena Biosciences. J.D.G. is founder, co‐owner, and member of the board of directors of Betascreen SL. J.D.G. is currently a full‐time employee of AstraZeneca. RV's institution has Clinical Research Agreements (RV as PI) with Alector, AviadoBio, Biogen, BMS, J&J, and UCB. R.V.’s institution has consultancy agreements (R.V. as DSMB chair or member) with AC Immune and Novartis. F.B. is a steering committee or data safety monitoring board member for Biogen, Merck, Eisai, and Prothena; advisory board member for Combinostics, Scottish Brain Sciences, and Alzheimer Europe; and consultant for Roche, Celltrion, Rewind Therapeutics, Merck, and Bracco. F.B. has research agreements with ADDI, Merck, Biogen, GE Healthcare, and Roche, and is co‐founder and shareholder of Queen Square Analytics LTD. E.S.L., Y.A., L.L., L.E.C., D.V.G., A.dB., M.B., P.G., N.VT., and I.C. have no disclosures. Author disclosures are available in the supporting information.
Figures

References
MeSH terms
Substances
Grants and funding
- No 115952/Innovative Medicines Initiative
- European Union's Horizon 2020 research
- innovation programme
- GE Healthcare
- Springer Healthcare
- #101108819/MSCA
- #23AARF-1029663/Alzheimer Association Research Fellowship
- MICIU/AEI/10.13039/501100011033/Spanish Research Agency
- RYC2022-038136-I/Spanish Research Agency
- PID2022-143106OA-I00/European Union FSE+
- European Union FEDER
- Alzheimer's Disease Data Initiative
- 23S06083-001
- Stichting Alzheimer Onderzoek
- Internal Funds KU Leuven
- G0G1519N/Flemish Research Foundation
- G094418N/Flemish Research Foundation
- HBC.2019.2523/VLAIO
- NIHR biomedical research centre
LinkOut - more resources
Full Text Sources
Medical

