This is a preprint.
Digital Twin for the Win: Personalized Cardiac Electrophysiology
- PMID: 40950065
- PMCID: PMC12424819
- DOI: 10.1101/2025.09.03.674034
Digital Twin for the Win: Personalized Cardiac Electrophysiology
Abstract
Background: Individual variability influences disease susceptibility, therapeutic response, and the emergence of rare phenotypes in both inherited and acquired cardiac diseases. Conventional preclinical models intentionally reduce variability to isolate biological effects but consequently fail to capture the heterogeneity present in human populations. This limitation contributes to translational gaps, incomplete mechanistic understanding, and adverse drug reactions. Digital twins offer a novel solution by integrating individualized data with simulation-based inference to predict physiology and therapeutic outcomes in a cell-specific, and ultimately, patient specific manner.
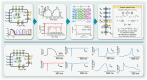
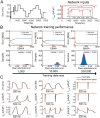
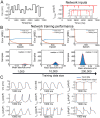
Methods: We developed an integrated computational, experimental, and machine learning framework to generate digital twins of human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs). A synthetic population of over one million computational iPSC-CMs was created by introducing physiologically plausible variation in 52 biophysical parameters governing six major ionic currents. Two synthetic datasets were used to train and test a fully connected deep neural network to infer complete parameter sets directly from optimized whole-cell voltage-clamp recordings.
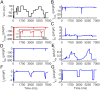
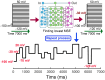
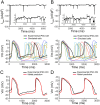
Results: Application of the model to experimental iPSC-CMs enabled rapid extraction of ionic conductance and kinetic parameters, generating digital twins that reproduced action potential waveforms and pacing frequencies. The models captured fine-scale electrophysiological features of depolarization, plateau, and repolarization with high fidelity across diverse morphologies and recording conditions. The framework was robust to both temperature perturbations and broad morphological variability, as we show that the synthetic training data can be readily re-tuned to any recording temperature and to include broad AP phenotypes.
Conclusions: This work introduces a scalable technology for generating fully parameterized, cell-specific digital twins of human iPSC-CMs from a single recording. By unifying computational modeling, synthetic data generation, and deep learning, the approach transforms a slow, multi-step process into a rapid, versatile platform for personalized diagnostics, targeted therapeutics, and predictive safety pharmacology.
Keywords: Artificial Intelligence; Cardiac Electrophysiology; Computational Biology; Digital twin; Precision Medicine; Stem Cell Biology; computational modeling and simulations; deep learning; iPSC-CM; personalized cardiac electrophysiology.
Figures

References
-
- Gintant G, Sager PT and Stockbridge N. Evolution of strategies to improve preclinical cardiac safety testing. Nat Rev Drug Discov. 2016;15:457–71. - PubMed
-
- Laverty H, Benson C, Cartwright E, Cross M, Garland C, Hammond T, Holloway C, McMahon N, Milligan J, Park B, Pirmohamed M, Pollard C, Radford J, Roome N, Sager P, Singh S, Suter T, Suter W, Trafford A, Volders P, Wallis R, Weaver R, York M and Valentin J. How can we improve our understanding of cardiovascular safety liabilities to develop safer medicines? Br J Pharmacol. 2011;163:675–93. - PMC - PubMed
-
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG, Inflammation and Host Response to Injury LSCRP. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–12. - PMC - PubMed
-
- Sorger PK and Allerheiligen SRB. Quantitative and Systems Pharmacology in the Post-genomic Era: New Approaches to Discovering Drugs and Understanding Therapeutic Mechanisms. Tech Rep. 2011.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
