This is a preprint.
Single-vessel transcriptome map pathological landscapes and reveal NR2F2-mediated smooth muscle cell phenotype acquisition in capillary malformations
- PMID: 40950147
- PMCID: PMC12424823
- DOI: 10.1101/2025.09.02.673874
Single-vessel transcriptome map pathological landscapes and reveal NR2F2-mediated smooth muscle cell phenotype acquisition in capillary malformations
Abstract
Background: Capillary malformation (CM) is a congenital vascular anomaly affecting the skin, mucosa, and brain, yet the understanding of its vascular pathogenesis remains limited.
Methods: We applied spatial whole-transcriptome profiling (GeoMx) and gene set enrichment analysis within CM lesions at single vasculature level. Differentially expressed genes were validated by immunofluorescence staining. Phosphoproteomics was profiled to uncover lesion-wide phosphorylation sites on proteins. Single-cell RNA sequencing was performed on CM-derived induced pluripotent stem cells (iPSCs) to determine differentiation trajectories of lesional vascular lineages. In silico gene perturbation was used to predict candidate genes for modulating vascular pathological progression, followed by functional validation in CM iPSC-derived endothelial cells (ECs) using a Tet-on system.
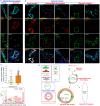
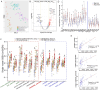
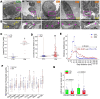
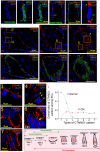
Results: A spatial transcriptomic atlas was constructed, and pathological landscape of individual CM vasculature was delineated. CM vessels exhibited hallmarks of endothelial-to-mesenchymal transition (EndMT), including disruption of adherens junctions (AJs), vascular identity transitions, and metabolic remodeling. Phosphoproteomics confirmed that differentially phosphorylated proteins were enriched in EndMT- and AJ-related pathways. Aberrant expression of venous transcriptional factor NR2F2 was observed in lesional ECs and correlated with progressive enlargement from capillaries to larger-caliber vessels containing multiple layers of smooth muscle cells (SMCs). In CM iPSCs, differentiation course yielded reduced ECs but increased SMCs. In silico knockout simulation predicted NR2F2 as a crucial regulator of facilitating SMC phenotype in CM. Consistently, enforced NR2F2 expression during iPSC differentiation suppressed endothelial markers while inducing SMC-associated genes.
Conclusions: Single CM vasculature displays pathological hallmarks characterized by EndMT and AJ disruption, leading to progressive vascular remodeling. NR2F2 functions as a central regulatory factor orchestrating the acquisition of the SMC phenotype, thereby representing a potential therapeutic target in CM.
Keywords: Capillary malformation; NR2F2; adherens junctions; endothelial cells; endothelial-to-mesenchymal transition; induced pluripotent stem cells; whole transcriptome atlas.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Eifert S, Villavicencio JL, Kao TC, Taute BM, Rich NM. Prevalence of deep venous anomalies in congenital vascular malformations of venous predominance. J Vasc Surg. 2000;31:462–471. - PubMed
-
- ISSVA. ISSVA Classification of Vascular Anomalies. International Society for the Study of Vascular Anomalies 2018;issva.org/classification.
-
- Lever WF, Schaumburg-Lever G. Histopathology of the Skin. 7th ed. Philadelphia, PA: J.B. Lippincott Co.; 1990.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
