This is a preprint.
Systematic analysis of snRNA genes reveals frequent RNU2-2 variants in dominant and recessive developmental and epileptic encephalopathies
- PMID: 40950445
- PMCID: PMC12424890
- DOI: 10.1101/2025.09.02.25334923
Systematic analysis of snRNA genes reveals frequent RNU2-2 variants in dominant and recessive developmental and epileptic encephalopathies
Abstract
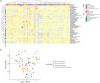
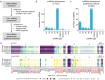
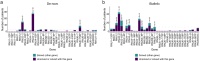
Variants in spliceosomal small nuclear RNA (snRNA) genes RNU4-2 (ReNU syndrome), RNU5B-1, and RNU2-2 have recently been linked to dominant neurodevelopmental disorders (NDDs), revealing a major, previously overlooked role for noncoding snRNAs in human disease. Here, we systematically analysed 200 potentially functional snRNA genes in a French cohort comprising 26,911 individuals with rare disorders and through international collaborations. We identify de novo and biallelic variants in RNU2-2 associated with both dominant and recessive NDDs in 126 individuals from 108 unrelated families. Recessive RNU2-2 NDD is at least twice as frequent as the dominant NDD caused by n.4G>A and n.35A>G, and often arises from a de novo variant in trans with an inherited allele, reflecting the high mutability of snRNA genes. Dominant and recessive RNU2-2-NDDs share overlapping clinical features with frequent epilepsy. Blood transcriptomics and DNA methylation analyses revealed subtle, variant-specific effects on splicing and episignatures. Our findings support a gradient-of-impact model and a continuum between dominant and recessive inheritance, establishing RNU2-2 variants as a frequent cause of NDDs, nearly as prevalent as ReNU syndrome.
Conflict of interest statement
N.W. receives research funding from Novo Nordisk and Biomarin Pharmaceutical. L.T.D. receives research funding from Stoke Therapeutics. L.G.S. receives funding from the Health Research Council of New Zealand and Cure Kids New Zealand. She has served as a paid consultant of the Epilepsy Study Consortium for consulting work for Epygenix Therapeutics, Ovid Therapeutics, Stoke Therapeutics, Takeda Pharmaceuticals, UCB, and Zogenix. L.G.S. has received research grants and consultancy fees from Zynerba Pharmaceuticals and has served on Takeda and Eisai Pharmaceuticals scientific advisory panels. I.E.S. has served on scientific advisory boards for CAMP4 Therapeutics, Longboard Pharmaceuticals, Mosaica Therapeutics, Takeda Pharmaceuticals, UCB; has received speaker honoraria from Akumentis, Biocodex, Chiesi, Stoke Therapeutics, UCB, Zuellig Pharma; has received funding for travel from Stoke Therapeutics and UCB; has served as an investigator for Anavex Life Sciences, Biohaven Ltd, Bright Minds Biosciences, Cerebral Therapeutics, Cerecin Inc, Cereval Therapeutics, Encoded Therapeutics, EpiMinder Inc, ES-Therapeutics, Longboard Pharmaceuticals, Marinus, Neuren Pharmaceuticals, Neurocrine BioSciences, Praxis Precision Medicines, Shanghai Zhimeng Biopharma, SK Life Science, Supernus Pharmaceuticals, Takeda Pharmaceuticals, UCB, Ultragenyx, Xenon Pharmaceuticals, Zogenix; and has consulted for Biohaven Pharmaceuticals, Eisai, Epilepsy Consortium, Longboard Pharmaceuticals, Praxis, Stoke Therapeutics, UCB; and is a Non-Executive Director of Bellberry Ltd and a Director of the Australian Academy of Health and Medical Sciences. She may accrue future revenue on pending patent WO61/010176 (filed: 2008): Therapeutic Compound; has a patent for SCN1A testing held by Bionomics Inc and licensed to various diagnostic companies; has a patent molecular diagnostic/theranostic target for benign familial infantile epilepsy (BFIE) [PRRT2] 2011904493 & 2012900190 and PCT/AU2012/001321 (TECH ID:2012-009). All other authors declare no competing interests.
Figures











References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
