Common food preservatives induce an oxidative stress response in Salmonella enterica serovar Typhimurium
- PMID: 40952897
- PMCID: PMC12440568
- DOI: 10.1099/mic.0.001609
Common food preservatives induce an oxidative stress response in Salmonella enterica serovar Typhimurium
Abstract
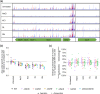
Despite their frequent use, the mechanisms of action of common food preservatives are poorly understood. As there is a drive to develop alternative preservatives, understanding the mechanisms of action of current preservatives can inform the development of novel food preservatives to ensure their efficacy. Here, we used TraDIS-Xpress, a large-scale, genome-wide unbiased screen to determine the mechanisms of action of common food preservatives by determining the genes that affect preservative susceptibility in Salmonella enterica serovar Typhimurium. We identified genes associated with central metabolism and oxidative stress responses that were important for all four preservatives. Formate dehydrogenase activity and synthesis was crucial for survival in the presence of both sodium chloride and potassium chloride. We found some preservative-specific effects on pathogen susceptibility, for example, LPS synthesis which improved survival upon exposure to sodium nitrite but harmed survival when exposed to sodium chloride or potassium chloride. This research expands our understanding of how some current preservatives act and can inform the effective use of preservatives in current and emerging food products to ensure high standards of food safety.
Keywords: functional genomics; TraDIS; transposon mutagenesis.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


References
-
- Henney J, Taylor C, Boon C. Strategies to Reduce Sodium Intake in the United States. Washington, DC: National Academies Press (US); 2010. Preservation and physical property roles of sodium in foods. - PubMed
-
- Albarracín W, Sánchez IC, Grau R, Barat JM. Salt in food processing; usage and reduction: a review. Int J of Food Sci Tech. 2011;46:1329–1336. doi: 10.1111/j.1365-2621.2010.02492.x. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

