De novo discovery of conserved gene clusters in microbial genomes with Spacedust
- PMID: 40954296
- PMCID: PMC12510874
- DOI: 10.1038/s41592-025-02816-x
De novo discovery of conserved gene clusters in microbial genomes with Spacedust
Abstract
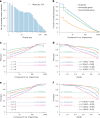
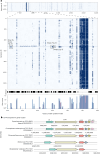
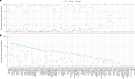
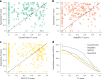
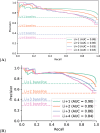
Metagenomics has revolutionized environmental and human-associated microbiome studies. However, the limited fraction of proteins with known biological processes and molecular functions presents a major bottleneck. In prokaryotes and viruses, evolution favors keeping genes participating in the same biological processes colocalized as conserved gene clusters. Conversely, conservation of gene neighborhood indicates functional association. Here we present Spacedust, a tool for systematic, de novo discovery of conserved gene clusters. To find homologous protein matches, Spacedust uses fast and sensitive structure comparison with Foldseek. Partially conserved clusters are detected using novel clustering and order conservation P values. We demonstrate Spacedust's sensitivity with an all-versus-all analysis of 1,308 bacterial genomes, identifying 72,843 conserved gene clusters containing 58% of the 4.2 million genes. It recovered 95% of antiviral defense system clusters annotated by the specialized tool PADLOC. Spacedust's high sensitivity and speed will facilitate the annotation of large numbers of sequenced bacterial, archaeal and viral genomes.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures








References
-
- Quince, C., Walker, A. W., Simpson, J. T., Loman, N. J. & Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol.35, 833–844 (2017). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

