Multi-Millennial Genetic Resilience of Baltic Diatom Populations Disturbed in the Past Centuries
- PMID: 40955567
- PMCID: PMC12439084
- DOI: 10.1111/gcb.70467
Multi-Millennial Genetic Resilience of Baltic Diatom Populations Disturbed in the Past Centuries
Abstract
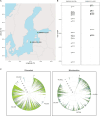
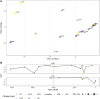
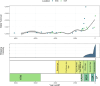
Little is known about the genetic diversity and stability of natural populations over millennial time scales, although the current biodiversity crisis calls for heightened understanding. Marine phytoplankton, the primary producers forming the basis of food webs in the oceans, play a pivotal role in maintaining marine ecosystems health and serve as indicators of environmental change. This study examines the genetic diversity and shifts in allelic composition in the diatom species Skeletonema marinoi over ~8000 years in the Baltic Sea by analyzing chloroplast and mitochondrial genomes. Sedimentary ancient DNA (sedaDNA) demonstrates the stability and resilience of genetic composition and diversity of this species across millennia in the context of major climate events. Accelerated change in allelic composition is observed from historical periods onwards, coinciding with times of intensifying human activity, like the Roman Empire, the Viking Age, and the Hanseatic Age, suggesting that anthropogenic stressors have profoundly impacted this species for the last two millennia. The data indicate a very high natural stability and resilience of the genomic composition of the species and underscore the importance of uncovering genomic disruptions caused by human impact on organisms, even those not directly exploited, to better predict and manage future biodiversity.
Keywords: Skeletonema marinoi; Baltic Sea; phytoplankton; population genomics; sedaDNA; target capture.
Global Change Biology© 2025 The Author(s). Global Change Biology published by John Wiley & Sons Ltd.
Conflict of interest statement
In the preparation of this manuscript, we employed artificial intelligence (AI) tools to enhance the quality of our work and to optimize our research process. R Script Optimization: we utilized an AI‐based code optimization to refine our R scripts. The tool provided suggestions for code enhancement, identified potential bugs, and recommended more efficient coding practices. This not only improved the performance of our scripts but also ensured the reproducibility and reliability of our results. We believe that the use of AI in our research process has improved the quality of our work. However, we stress that the final decisions on manuscript content and the interpretation of the results were made by the authors.
The authors declare no conflicts of interest.
Figures

References
-
- Andrén, T. , Björck S., Andrén E., Conley D., Zillén L., and Anjar J.. 2011. “The Development of the Baltic Sea Basin During the Last 130 Ka.” In The Baltic Sea Basin, edited by Harff J., Björck S., and Hoth P., 75–97. Springer. 10.1007/978-3-642-17220-5_4. - DOI
-
- Andrews, S. 2021. “FastQC: A Quality Control Analysis Tool for High Throughput Sequencing Data [Java].” https://github.com/s‐andrews/FastQC.
-
- Barbier, E. B. , ed. 2010. “The Contemporary Era (From 1950 to the Present).” In Scarcity and Frontiers: How Economies Have Developed Through Natural Resource Exploitation, 552–662. Cambridge University Press. 10.1017/CBO9780511781131.010. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

